Figure 6.
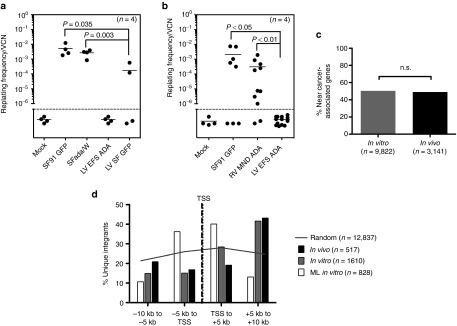
Assessments of genotoxicity of EFS-ADA lentiviral vector. In vitro immortalization (IVIM) Assay. Murine Lin- cells transduced by the indicated vectors were expanded as mass cultures for 2 weeks. An aliquot was taken for qPCR for VCN measurement. On day 15, cells were plated into 96-well plates at a density of 100 cells/well or 1,000 cells/well in 100 µl medium. Two weeks later, the wells showing abundant cell growth were counted as positive, and the frequency of replating cells was calculated based on Poisson statistics using L-Calc Software (Stemcell Technologies, Vancouver, Canada). Horizontal bars indicate mean values. (a) Replating frequency corrected for VCN group by investigators at GOSH, UK. (b) Replating frequency corrected for VCN group by investigators at University of California, Los Angeles, USA (c,d) Vector integration site analysis in human ADA-deficient bone marrow in vitro and in vivo. (c) The percentages of unique integration sites in human cells (isolated from primary NSG mouse recipient bone marrow) near cancer-related genes were determined in vitro (n = 9,822 unique sites) or in vivo (n = 3,141 unique sites). Integration sites in genes or within 300 kb of gene TSS were considered “near” and cancer-related genes were defined as in Higgins et al. 40.(d) The EFS-ADA vector integration sites were mapped relative to transcriptional start sites (TSS) in vitro (n = 1,610 unique sites) and in vivo (n = 517), and compared to a published data set for MLV1 (n = 828). Grey line represents the theoretical random distribution (n = 12,837).
