Figure 1.
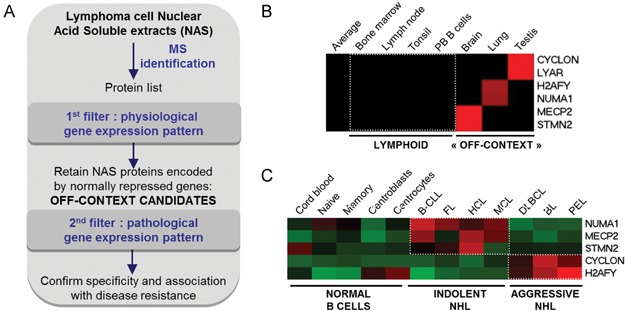
- Schematic representation of the overall experimental strategy, as indicated. NAS: nuclear acid-soluble, MS: mass spectrometry.
- Heat-map representation of the gene expression levels for 6 candidate ‘off-context’ genes in normal lymphoid and non-lymphoid tissues; PB: peripheral blood [data from BioGPS (Su et al, 2004)].
- Heat-map representation of gene expression profiles for 5 ‘off-context’ genes, as indicated; NHL: non-Hodgkin lymphoma, B-CLL: B-cell chronic lymphocytic leukemia, FL: follicular lymphoma, HCL: hairy cell leukemia, MCL: mantle cell lymphoma, BL: Burkitt lymphoma, DLBCL: diffuse large B-cell lymphoma, PEL: primary effusion lymphoma [data from GEO GSE2350 (Basso et al, 2005)].
