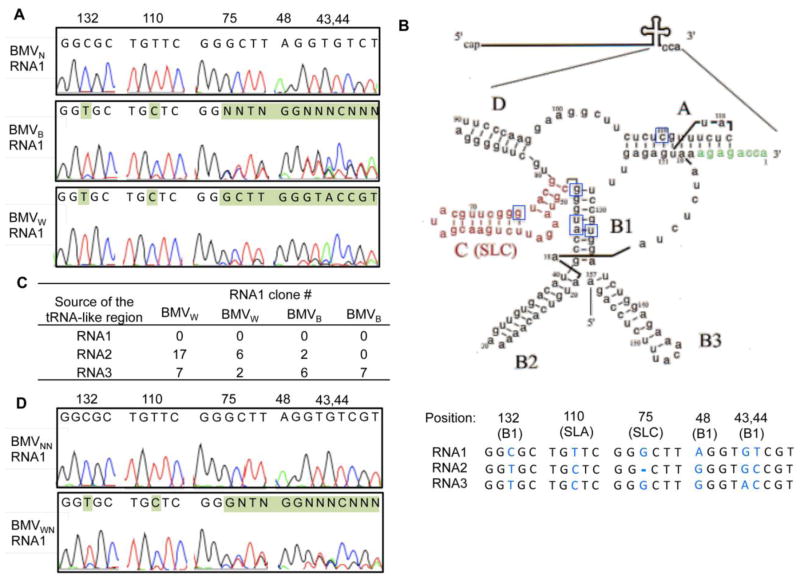
Fig. 2. RNA recombination occurred within the tRNA-like structure of RNA1 in BMVW and BMVB.
A) Representative sequencing chromatograms of the cDNA population derived from the tRNA-like structure in RNA1 for BMVW, BMVB and BMVN. Residues shaded in green identified mismatches to the sequences present in the Agrobacterium constructs used to start BMV infection. The numbers indicate the position of the nucleotides, with the 3′-most nucleotide being residue 1. B) Schematic of the common tRNA-like structure and the sequences of the diagnostic sites. The schematic adapted from Felden et al. is modeled after the tRNA-like region in RNA3.67 The name of each stem-loop structure is in bold and the core promoter for minus-strand RNA synthesis, SLC, is in red. The nucleotides affected by the recombination in RNA1 are boxed. In the lower panel, the SNPs (nt 43, 44, 132) from the three BMV RNAs are colored blue with their positions shown above. In addition to the three SNPs, substitutions at nt 48 and 110 in RNA1 and a deletion at nt 75 in RNA2 are present in the BMV cDNA in the Agrobacterium. These five positions are used to map the intersegment crossovers in the RNA1 recombinants. C) The number of clones obtained from the RNA1 cDNA library of BMVW or BMVB that contained sequences mapped to the tRNA-like structure in RNA1, RNA2 or RNA3. Two independent preparations of BMVw and BMVB were used to generate the results. D) Representative sequencing chromatograms of the cDNA population derived from the tRNA-like structure in the RNA1 after BMVW and BMVN were passed through N. benthamiana. The progeny virions are denoted as BMVWN and BMVNN respectively.