Table 3.
Comparison of LD and HD models for gene expression data with L2 (top) and L1 (bottom) penalties
| Predictors |
|||||
|---|---|---|---|---|---|
| Model type | Overall | Selected | Log-likelihood | c-Statistic | 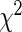 |
| Cox | |||||
| LD | 5 | 5 | −71.13 | 0.71 | 26.35 |
| HD | 24 496 | 24 026 | −72.10 | 0.76 | 12.75 |
| Exponential | |||||
| LD | 5 | 5 | −86.26 | 0.71 | 16.83 |
| HD | 24 496 | 24 344 | −98.72 | 0.75 | 112.69 |
| Weibull | |||||
| LD | 5 | 5 | −85.64 | 0.71 | 12.79 |
| HD | 24 496 | 22 299 | −85.56 | 0.70 | 9.34 |
| Log-logistic | |||||
| LD | 5 | 5 | −85.65 | 0.70 | 14.74 |
| HD | 24 496 | 22 090 | −86.14 | 0.70 | 5.37 |
| Log-normal | |||||
| LD | 5 | 5 | −52.10 | 0.70 | 16.02 |
| HD | 24 496 | 24 154 | −65.14 | 0.66 | 23.61 |
| Cox | |||||
| LD | 5 | 4 | −71.10 | 0.71 | 30.19 |
| HD | 24 496 | 20 | −72.64 | 0.71 | 7.49 |
| Exponential | |||||
| LD | 5 | 5 | −86.27 | 0.71 | 17.14 |
| HD | 24 496 | 13 | −87.51 | 0.67 | 2.74 |
| Weibull | |||||
| LD | 5 | 5 | −85.63 | 0.71 | 12.79 |
| HD | 24 496 | 13 | −86.80 | 0.66 | 3.75 |
| Log-logistic | |||||
| LD | 5 | 5 | −85.65 | 0.70 | 14.71 |
| HD | 24 496 | 9 | −86.20 | 0.68 | 3.66 |
| Log-normal | |||||
| LD | 5 | 5 | −52.10 | 0.70 | 15.98 |
| HD | 24 496 | 9 | −53.46 | 0.66 | 8.57 |
For L2 penalization, the number of selected predictors refers to the number of significant predictors 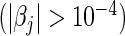 . In both cases, bold emphases represent superior performance of one type of Cox model over the other. For a more comprehensive comparison, the results obtained using four parametric models (Mittal and others, 2013) are also presented in the lower parts of the table.
. In both cases, bold emphases represent superior performance of one type of Cox model over the other. For a more comprehensive comparison, the results obtained using four parametric models (Mittal and others, 2013) are also presented in the lower parts of the table.
