Figure 5. Rapid kinetics of ligand binding to the trypsin-like proteases thrombin, FXa, and aPC.
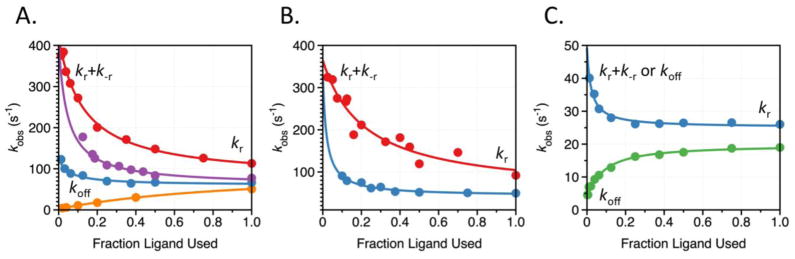
Rates of approach to equilibrium, kobs, determined from stopped-flow fluorescence measurements of binding of PABA (red), KCl (magenta), NaCl (blue), VPR (orange), and CaCl2 to (A) thrombin, (B) FXa, and (C) aPC. Reaction conditions were as follows: (A) 5 mM Tris, 0.1% PEG8000, pH 8.0 at 15 °C, with ionic strength kept constant at 400 mM with ChCl; (B) 50 mM Tris, 0.1% PEG8000, pH 8.0 at 15 °C, with no ChCl used due to specific inhibition of FXa by choline; (C) 5 mM Tris, 0.1% PEG8000, pH 8.0 at 15 °C, with ionic strength kept constant at 800 mM with ChCl. Solid lines were drawn according to the equation kr+k−r/{1+[L]/Kd} for PABA and KCl in thrombin, PABA and NaCl in FXa, and NaCl in aPC, or according to eq. 2 in the text. Best-fit parameters for thrombin binding to ligand (A) are: (PABA) kr=70±10 s−1, k−r=340±10 s−1, Kd=52±8 μM; (K+) kr=60±10 s−1, k−r=340±10 s−1, Kd=19±2 mM; (Na+) kr=60±10 s−1, k−r=340±10 s−1, kon=3.2±0.3 104 M−1s−1, koff=130±20 s−1; (VPR) kr=80±10 s−1, k−r=340±10 s−1, kon=1.7±0.1 107 M−1s−1, koff=2.4±0.2 s−1. Best-fit parameters for FXa binding to ligand (B) are: (PABA) kr=48±36 s−1, k−r=327±29 s−1, Kd=44±2 μM; (Na+) kr=43±4 s−1, k−r=240±20 s−1, Kd=10±1 mM. Best-fit parameters for aPC binding to ligand (C) are: (Na+) kr=25±1 s−1, k−r=24±4 s−1, Kd=9±1 mM; (Ca2+) kr=20±1 s−1, k−r=25±4 s−1, kon=16±1 103 M−1s−1, koff=5.0±0.3 s−1. Because k−r was not well determined in FXa binding to Na+, it was restricted by the values derived from PABA binding. A similar procedure was used for kon for Ca2+ binding to aPC. To facilitate comparison, the values of [L] for each ligand are expressed as the fraction of maximal ligand used.
