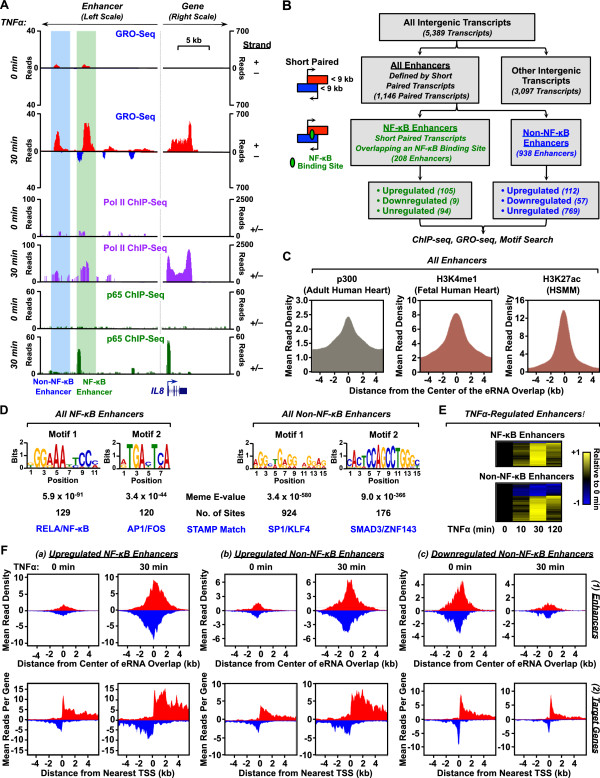
Figure 6.
Enhancer transcripts in AC16 cells originate from NF-κB-dependent and NF-κB-independent genomic loci. A) Genome browser tracks showing read distributions for GRO-seq, Pol II ChIP-seq, and p65 ChIP-seq at the promoter and distal enhancers of the IL8 gene. The blue-shaded genomic region shows an NF-κB-independent enhancer, whereas the green-shaded genomic region shows a NF-κB-dependent enhancer. A schematic of the IL8 gene annotation is shown and the length scale is indicated. B) Flowchart of enhancer classification in AC16 cells based on genomic location, eRNA production, length of the transcribed regions, overlap with NF-κB binding, and TNFα-mediated regulation. C) Metagene representations of the average ChIP-seq read distributions for p300 in adult human heart (left), H3K4me in fetal human heart (middle), and H3K27ac in human skeletal muscle myoblasts (HSMM) (right) for all 1,146 enhancers identified by GRO-seq, shown relative to the midpoint of overlap of the bidirectionally transcribed eRNAs (± 4 kb). D) De novo motif analyses of 208 NF-κB-dependent enhancers (left) and 938 NF-κB-independent enhancers (right) using MEME/STAMP. The top two most enriched motifs for each category are shown. E) Heatmap representations of TNFα regulation of enhancer transcription for NF-κB-dependent (top) and NF-κB-independent (bottom) enhancers. The color-based scale represents GRO-seq reads at the indicated time points scaled to the read density at time zero. F) Metagene representations of the average GRO-seq read distributions ± 4 kb around (1) the midpoint of overlap of the bidirectionally transcribed eRNAs (top row) or (2) the TSSs of the nearest neighboring protein-coding or lncRNA putative target genes (bottom row) for the following groups of enhancers: (a) TNFα-upregulated NF-κB-dependent enhancers (left), (b) TNFα-upregulated NF-κB-independent enhancers (middle), and (c) TNFα-downregulated NF-κB-independent enhancers (right).