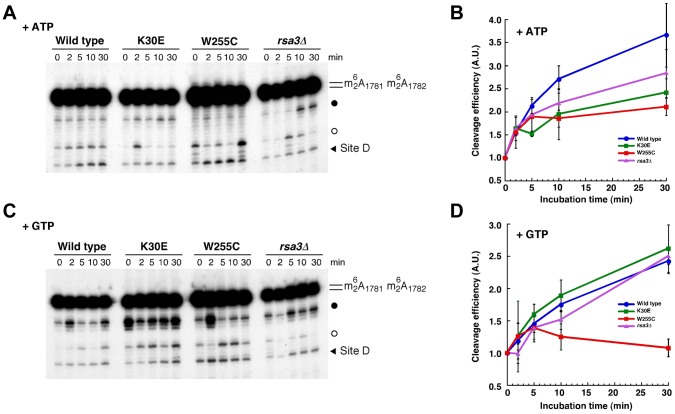
Figure 7. In vitro processing of 20S pre-rRNA is impaired in the rpl3[W255C] mutant.
In vitro cleavage assays were performed with pre-ribosomal particles purified via PTH-tagged Nob1 from different strains: wild-type (blue, circle), rpl3[W255C] (red, square), rpl3[K30E] (green, square) and rsa3Δ (purple, triangle). Purified particles were incubated in reaction buffer containing 1 mM ATP (A and B) or 1 mM GTP (C and D) for the indicated times (0, 2, 5, 10 and 30 min). RNA was extracted and cleavage at site D was analysed by primer extension with probe c' (Figure S1 and Table S3). Representative primer extension analyses are shown (A and C). The strong upper stops result from termination at sites of 18S rRNA base-dimethylation at A1781 and A1782. These modifications precede site D cleavage in vivo. The black arrow indicates site D. Filled and empty dots indicate non-relevant primer extensions stops that were observed in some experiments (for further discussion, see [26]). Signal intensities were measured by phosphoimager scanning; values were corrected for RNA loading using the dimethylation signals as internal standards, normalised to the sample at the zero time-point, arbitrarily set at 1.0, and plotted (B and D). The average of 2 (B) and 4 (D) independent experiments is shown; the error bars indicate the standard deviation.