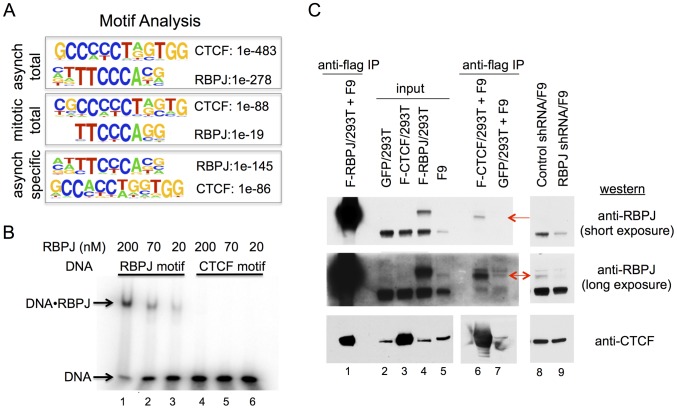
Figure 5. RBPJ interacts with CTCF and is enriched at genomic loci containing CTCF-binding motifs.
(A) Protein-binding motifs enriched at sites of RBPJ occupancy on chromatin from asynchronous cells and mitotic cells, as well as occupancy sites unique to asynchronous cells. The enrichment p-values for each motif are as shown. No significant motif was enriched at sites of RBPJ occupancy specific to mitotic chromatin. (B) Electrophoretic mobility shift assays showing binding of RBPJ to DNA containing an RBPJ-binding motif but not to DNA containing a CTCF-binding motif. 16 bp DNA fragments were used at 1 nM and RBPJ protein concentrations are as indicated. (C) Protein interaction experiments revealing RBPJ and CTCF association. Lysates prepared from 293T cells expressing Flag-RBPJ, Flag-CTCF or GFP were incubated with anti-Flag M2 agarose beads (α-Flag). After protein binding, the beads were washed and then incubated with F9 cell lysate. Bound proteins were eluted with Laemmli buffer and resolved in a NuPAGE 7% Tris-acetate gel (lanes 1, 6 and 7). RBPJ and CTCF were detected by western blot analysis, using antibodies indicated to the right. For the CTCF western, lanes 1–5 were from a long exposure and lanes 6–7 were from a short exposure. Lanes 2 and 5 are input lysates. Lanes 8 and 9 are lysates prepared from F9 cells expressing a control shRNA or an shRNA targeting RBPJ, revealing the presence of different RBPJ isoforms in F9 cells. CTCF was used as a loading control. Arrows point to the low abundance RBPJ isoform.