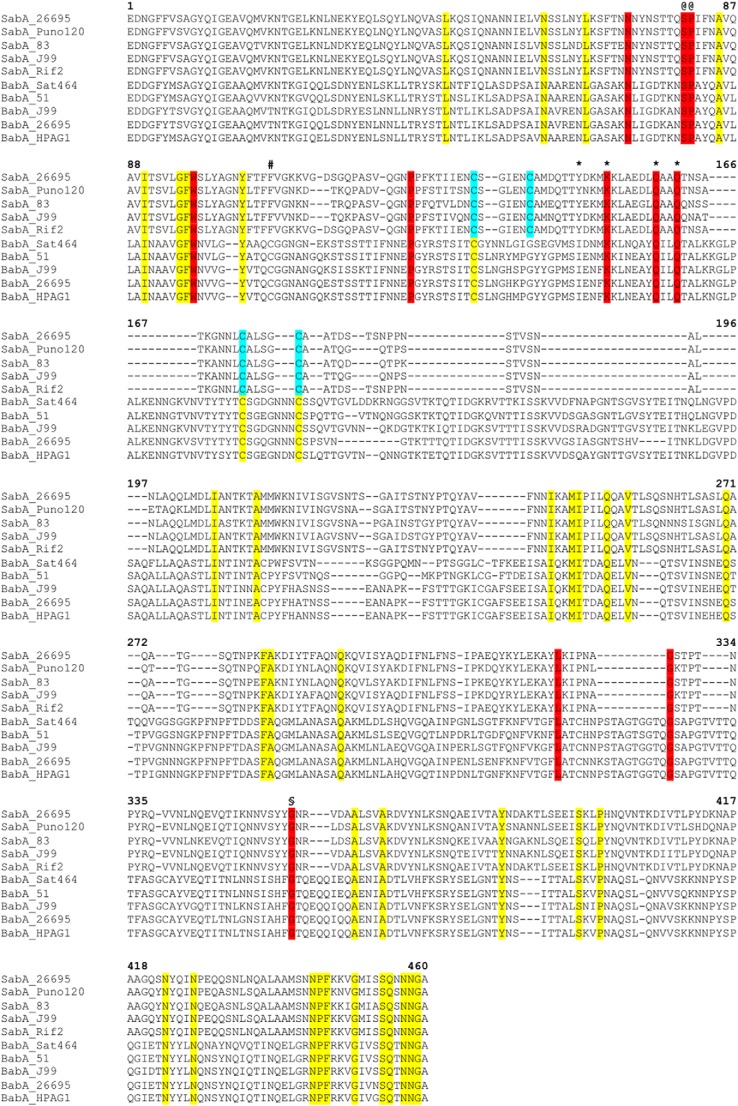
FIGURE 3.
Primary sequence alignment of SabA and BabA N-terminal adhesion regions. The alignment was generated with Clustal Omega using 33 SabA and 41 BabA sequences from various H. pylori strains (KEGG database). The figure shows a representative of the complete alignment (five sequences from each protein). Residues in yellow are identical in all SabA and BabA, red are identical surface residues in the predicted binding pocket, cyan are cysteine residues involved in disulfide bonds in SabA. The position of the alternative conserved cysteine residue in BabA is marked (#). Ser-80 and Pro-81 (@) map to the interruption in Helix-1, and Gly-357 (§) causes a slight kink in Helix-4. Tyr-148, Lys-152, Gln-159, and Gln-162 were mutated to alanine in this study are marked with an asterisk.