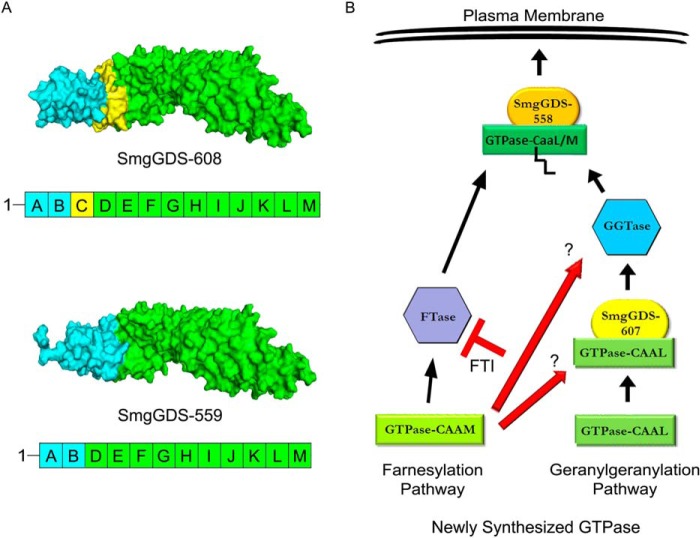
FIGURE 6.
SmgGDS-608/607 and SmgGDS-559/558 share similar structural domains with differences in the N-terminal region. A, PyMOL homology models of SmgGDS-608 and SmgGDS-559 are presented based on a previous published model of SmgGDS-608 (28). SmgGDS-608 corresponds to SmgGDS-607, whereas SmgGDS-559 corresponds to SmgGDS-558, as described under “Experimental Procedures.” The ARM domains in the proteins are designated A–M, with ARM C (yellow) being spliced out of SmgGDS-559/558. Both SmgGDS-608/607 and SmgGDS-559/558 have similar structures in ARMs D-M (green), but SmgGDS-608/607 differs significantly from SmgGDS-559/558 in the structure of ARMs A and B (blue). B, our results support a model in which a GTPase ending with a methionine enters the farnesylation pathway without the aid of SmgGDS-607. The farnesylated GTPase then interacts with SmgGDS-558 and traffics to the plasma membrane. In contrast, a small GTPase ending with a leucine interacts with SmgGDS-607 prior to becoming prenylated by the geranylgeranyltransferase. The geranylgeranylated GTPase then interacts with SmgGDS-558 and traffics to the plasma membrane. A FTI promotes the alternate geranylgeranylation of a GTPase that is normally farnesylated (e.g., K-Ras), and this leads to an increased association between a GTPase ending in a methionine and SmgGDS-607. More analyses are needed to determine whether the interaction of WT K-Ras with SmgGDS-607 is needed for the alternate geranylgeranylation of this GTPase in cells treated with FTI (red arrows).