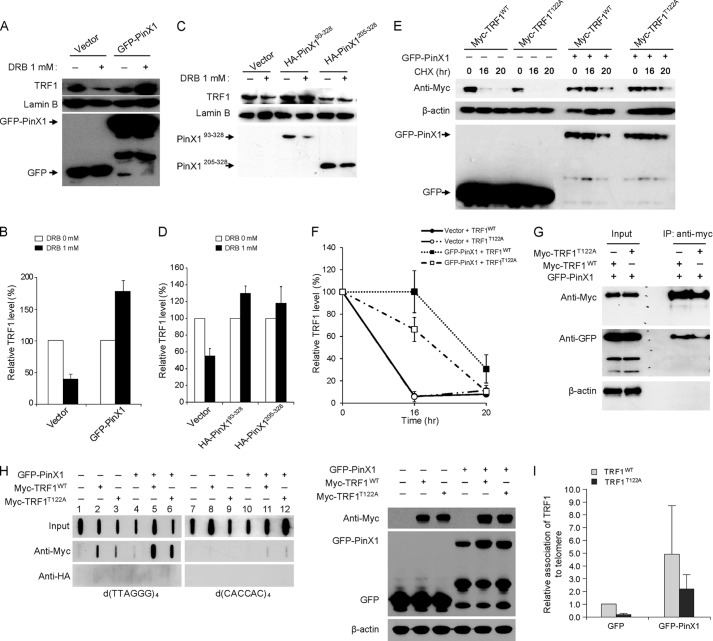
FIGURE 3.
PinX1 stabilizes TRF1T122A and allows the protein to bind to telomeres. A, increased stability of TRF1 in PinX1-expressing cells after treatment with DRB. HeLa cells transfected with GFP-PinX1 for 24 h were treated with 1 mm DRB for 8 h, and then nuclear lysates were resolved by SDS-PAGE, and proteins were detected by immunoblotting. B, graphical representation of relative TRF1 levels in A. The amount of TRF1 protein in DRB-treated cells was determined relative to those in DRB-untreated cells. Lamin B was used as a loading control, and error bars indicate S.D. from three independent experiments. C, PinX1 mutants containing TID domain also enhancing TRF1 stability. Nuclear lysates prepared from HeLa cells expressing HA-PinX1 constructs treated with DRB were resolved by SDS-PAGE, and proteins were detected by immunoblotting. D, quantification of data represented in C. The relative TRF1 levels were determined as described in B. Error bars indicate the S.D. from three independent experiments. E, PinX1 stabilizing TRF1T122A. HeLa cells co-transfected with GFP-PinX1 and myc-TRF1WT or myc-TRF1T122A were subjected to protein stability assays, and TRF1 was detected by anti-myc antibody. GFP vector was used as a control (−). F, quantification of data represented in E. TRF1 level was semiquantified using β-actin as a loading control, and the TRF1 amount at 0-h time point was defined as 100%. Error bars represent the S.D. of the mean from three independent experiments. G, interaction of PinX1 with TRF1T122A. HeLa cells expressing GFP-PinX1 and myc-TRF1WT or myc-TRF1T122A were subjected to immunoprecipitation with anti-myc antibody, followed by immunoblot with anti-myc and anti-GFP antibodies. H, increased TRF1T122A on telomeres in PinX1-overexpressed cells. HeLa cells co-transfected with GFP-PinX1 and myc-TRF1WT or myc-TRF1T122A were subjected to ChIP with anti-myc antibody. The presence of telomeres in the precipitated DNA was detected by hybridization with d(TTAGGG)4. HA ChIPs were performed as a negative control. Right panel shows immunoblots of the samples used in the ChIP assay. Expressions of GFP-PinX1, myc-TRF1WT, and myc-TRF1T122A were confirmed by immunoblotting. I, quantification of ChIPs in H. Telomere signals of myc ChIPs were normalized with input signals and calculated as a ratio relative to that recovered from myc-TRF1WT in GFP vector-expressing cells. Error bars indicate the S.D. from three independent experiments.