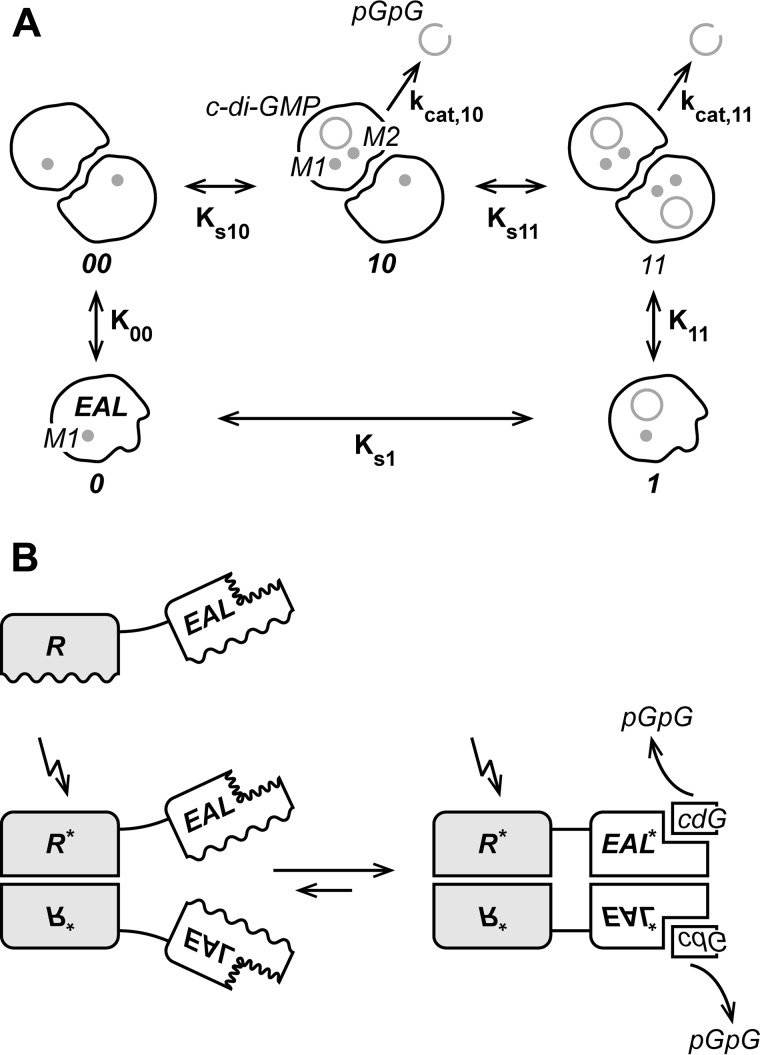
FIGURE 9.
A, thermodynamic scheme showing five YahA-EAL states that are in fast thermodynamic equilibrium: monomeric YahA-EAL in the uncomplexed (0) and cdG (ring symbol) complexed (1) state as well as dimeric YahA-EAL in the uncomplexed (00) and singly (10) or doubly (11) occupied dimeric state. Magnesium ions in sites M1 and M2 are indicated. The second-order ligand (Ks1, Ks10, and Ks11) and dimer (K00, K11) association constants and turnover numbers (kcat,10 and kcat,11) are indicated. Assuming no cooperativity, Ks10 = Ks11 and kcat,10 = kcat,11. Monomeric YahA-EAL is inactive, due to the postulated absence of a divalent cation in site M2. Dimeric YahA-EAL hydrolyzes cdG to yield the linear pGpG dinucleotide (open ring symbol). B, generic regulatory mechanism for a full-length EAL phosphodiesterase with associated regulatory domain (R). Wavey lines indicate interfaces that undergo structural changes. The protein is monomeric (top), but dimerizes via the R domains upon signal perception (bottom left). This promotes dimerization of the EAL domains, due to the increase of their local concentration (bottom left). Finally, structural changes in the EAL/EAL interface induced by dimerization are coupled to structural changes in the active site that would affect substrate affinity and/or catalytic activity.