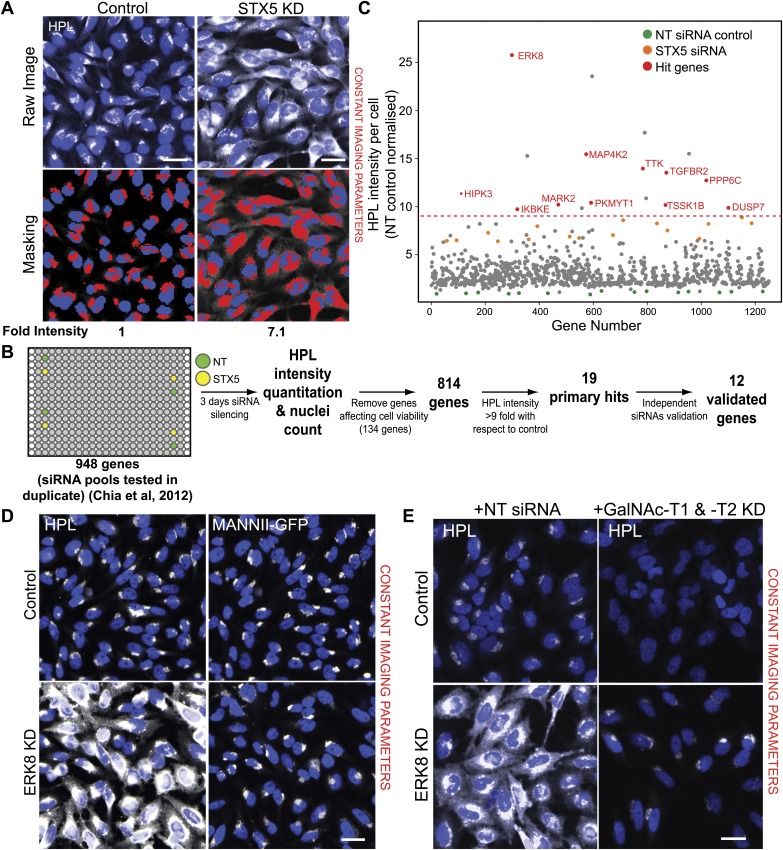
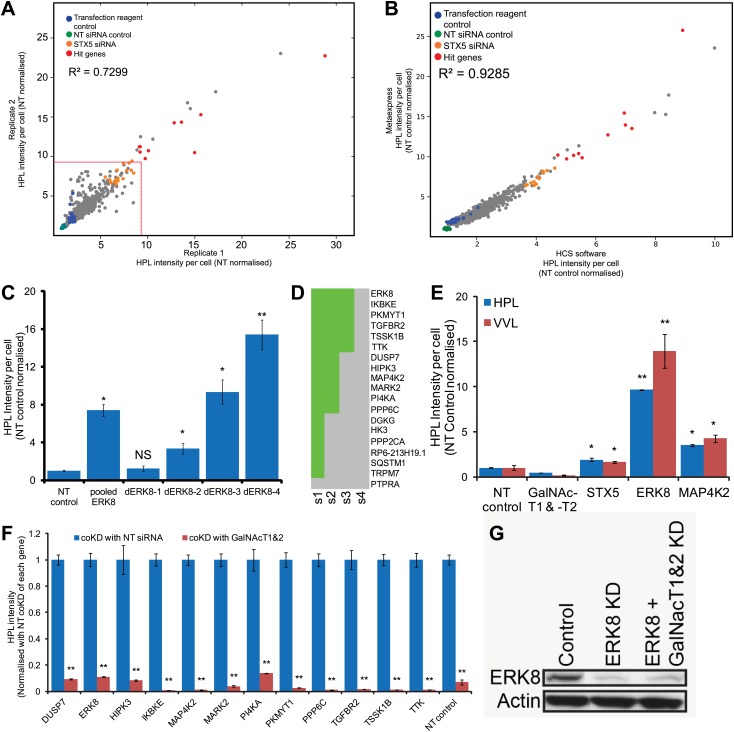
Figure 1. RNAi screening reveals 12 negative regulators of Tn expression.
(A) Helix pomatia lectin (HPL) staining was analysed using the ‘Transfluor HT’ module of MetaXpress software (Molecular Devices). A mask was generated for both HPL and nuclei (Hoechst) staining to classify the region of measurement (lower panels). Scale bar: 30 µm. (B) Schematic overview of the screening process. Images from the RNAi screen in Chia et al. (2012) were quantified for HPL intensities. Non-targeting (NT) siRNA and Syntaxin 5 (STX5) siRNA were used as negative and positive controls, resepectively. (C) Fold-change of HPL intensities normalised to NT siRNA treatment (green dots) and STX5 (orange dots). Primary hits were selected based on a threshold of a nine-fold increase (red dashed line) and the final validated genes are labelled in red (Hit genes). (D) Images from the screen of HPL staining in HeLa cells depleted of ERK8. MannII-GFP labels the Golgi apparatus. Scale bar: 30 μm. (E) HPL staining in cells knockdown of ERK8 with a control siRNA or GalNAc-T1 and -T2 siRNA. Scale bar: 30 μm.