Figure 1.
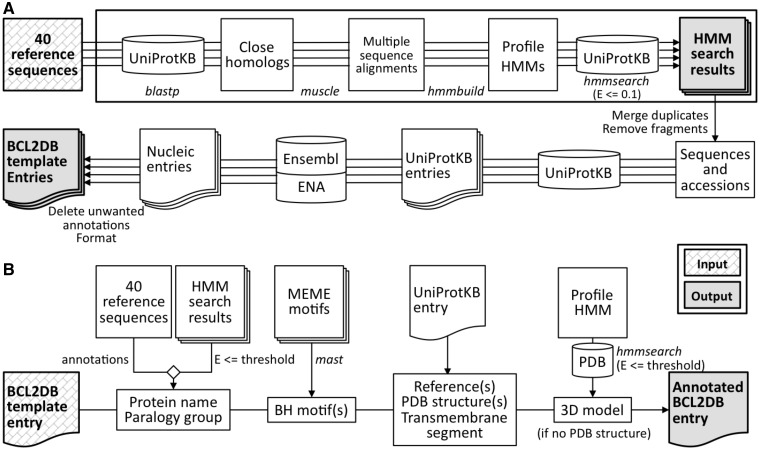
Description of the FindBCL2 and AnnotateBCL2 processes used to generate BCL2DB. External programs used by the processes are indicated in italics. (A) The upper part of the panel (boxed) describes the discovery mode of the FindBCL2 program. The results are the profile HMMs and their associated classification E-value thresholds deduced after a HMM search against UniProtKB. The production mode used to generate the BCL2DB entry templates is described in the bottom part. After an hmmsearch on UniProtKB with the computed profile HMMs, the Ensembl or ENA entries are retrieved from cross-references or BLAST searches with nonfragment protein sequences and after removing duplicated sequences. Then, the entries are cleared of unwanted annotations and merged into a single one if they refer to the same Ensembl, Ensembl Genomes or ENA entry. (B) The AnnotateBCL2 process enriches each BCL2DB entry template with annotations from reference sequences, sequence classification information (protein/gene name and orthology group/cluster), location of BH motifs and structural data retrieved from the PDB.