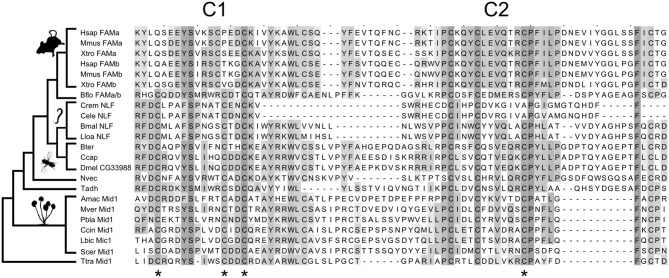
Figure 1.
Evolution of the Mid1 superfamily. Key cysteine residues are conserved between fungal and animal Mid1. Stars are placed under four cysteine residues that are necessary for Mid1 function in yeast (Maruoka et al., 2002). Three of the four are conserved in fungi, vertebrates and Caenorhabditis, and all four are conserved in all other sampled animals. The sampling of the key outgroups to vertebrates (Branchiostoma floridae) and Caenorhabditis (Brugia malayi and Loa loa) allows us to reconstruct the loss of key cysteines and confidently assign homology. The species sampled, from top to bottom are: Vertebrates: Hsap, Homo sapiens; Mmus, Mus musculus; Xtro, Xenopus tropicalis. Chordates: Bflo, Branchiostoma floridae. Nematodes: Crem, Caenorhabditis remanei; Cele, Caenorhabditis elegans; Bmal, Brugia malayi; Lloa, Loa loa. Insects: Bter, Bombus terrestris; Ccap, Ceratitis capitata; Dmel, Drosophila melanogaster. Basal Animals: Nvec, Nematostella vectensis; Tadh, Trichoplax adhaerens. Fungi: Amac, Allomyces macrogynus; Mver, Mortierella verticillata; Pbla, Phycomyces blakesleeanus; Ccin, Coprinopsis cinerea okayama; Lbic, Laccaria bicolor; Scer, Saccharomyces cerevisiae. Apusozoa: Ttra, Thecamonas trahens. The alignment is shaded by conservation, according to BLOSUM 64 matrix, and has had majority-gapped columns removed. Thumbnails were obtained from PhyloPic.org.