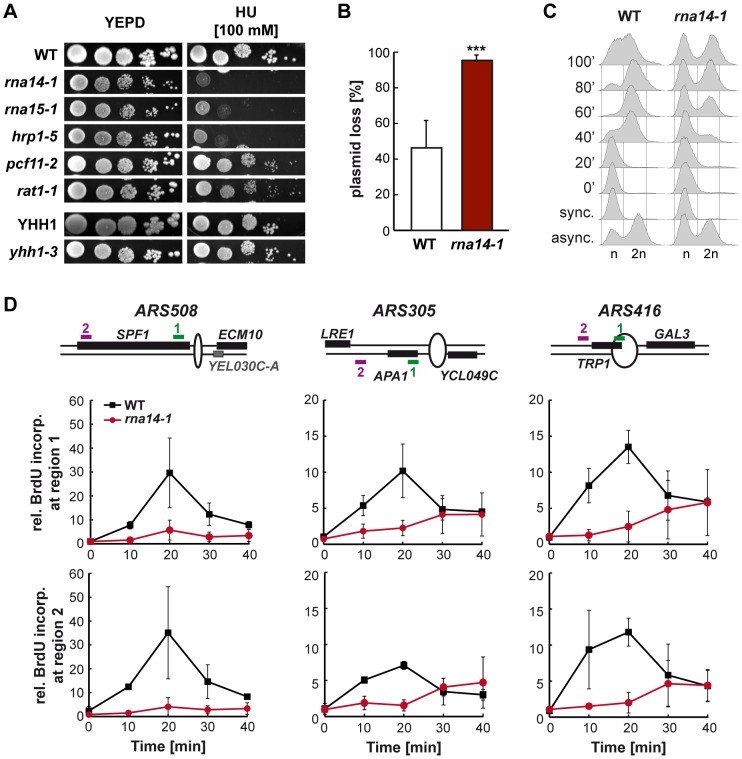
Figure 4. Cell cycle progression is compromised in rna14-1 cells.
(A) Sensitivity to hydroxyurea (HU) of six transcription-termination mutants. Serial dilutions (10-fold) of exponentially growing cultures are shown. (B) Analysis of plasmid loss in rna14-1, monitored as the percentage of cells that lost the pRS315 centromeric plasmid after ∼10 divisions in non-selective media. Average and standard deviation of six independent transformants are plotted for each genotype. Statistical analysis was performed with a two-tailed unpaired student t-test compared with the wild type. ***p<0.001. (C) Cell cycle progression analysis in wild-type (WT) and rna14-1 strains. Asynchronous (async.), α-factor synchronized (sync.) and released cells were analysed by FACS. (D) Analysis of replication in rna14-1 cells. BrdU incorporation upon release of G1-arrested cells was analysed at early replicating origins ARS508, ARS305, and ARS416 by immunoprecipitation and RT qPCR. A schematic drawing of each ARS and localization of the amplified regions are depicted (top). Quantification of BrdU incorporation relative to a late replicating locus is plotted for each region. Average from two independent experiments and corresponding standard deviations are shown.