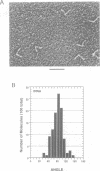
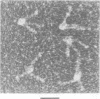
Abstract
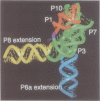
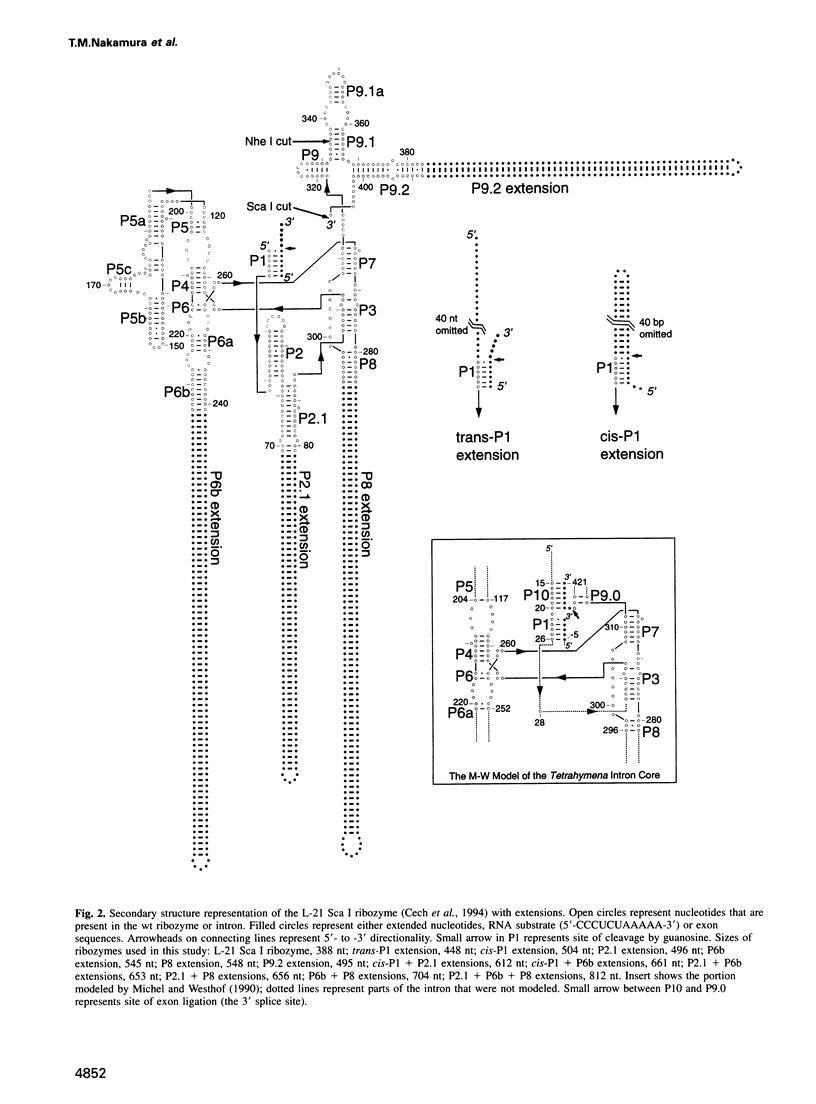
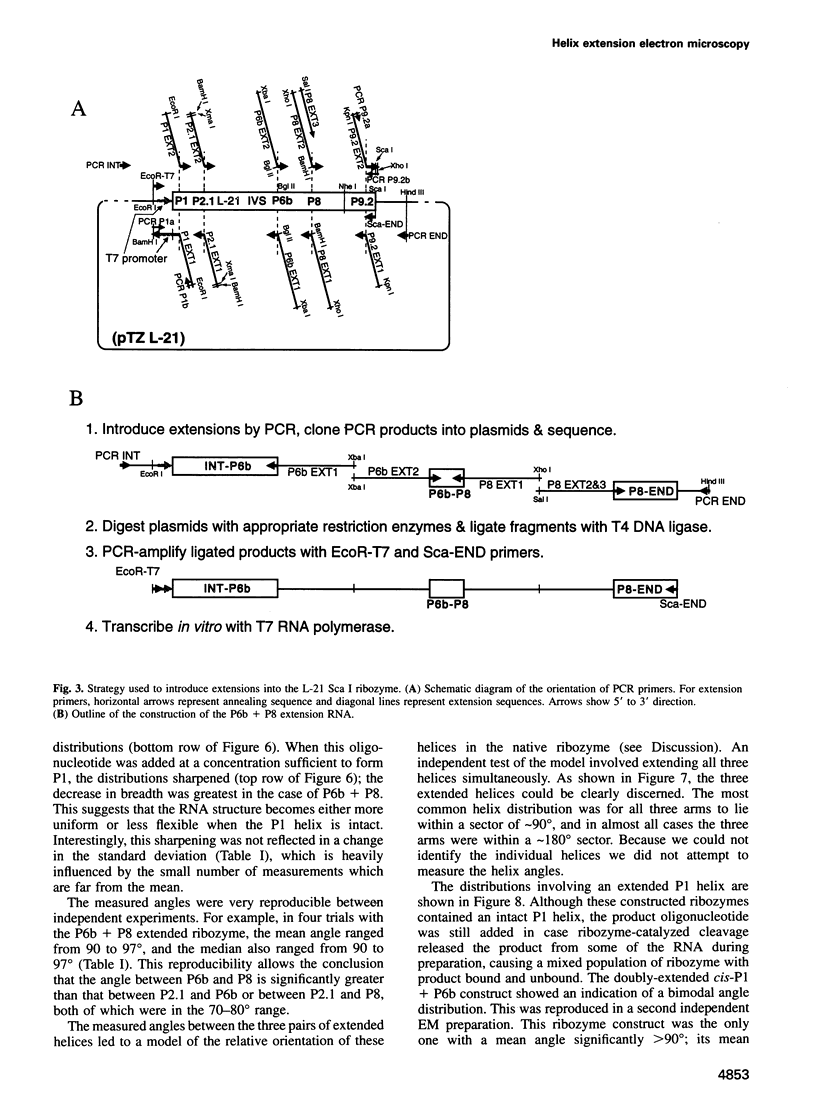
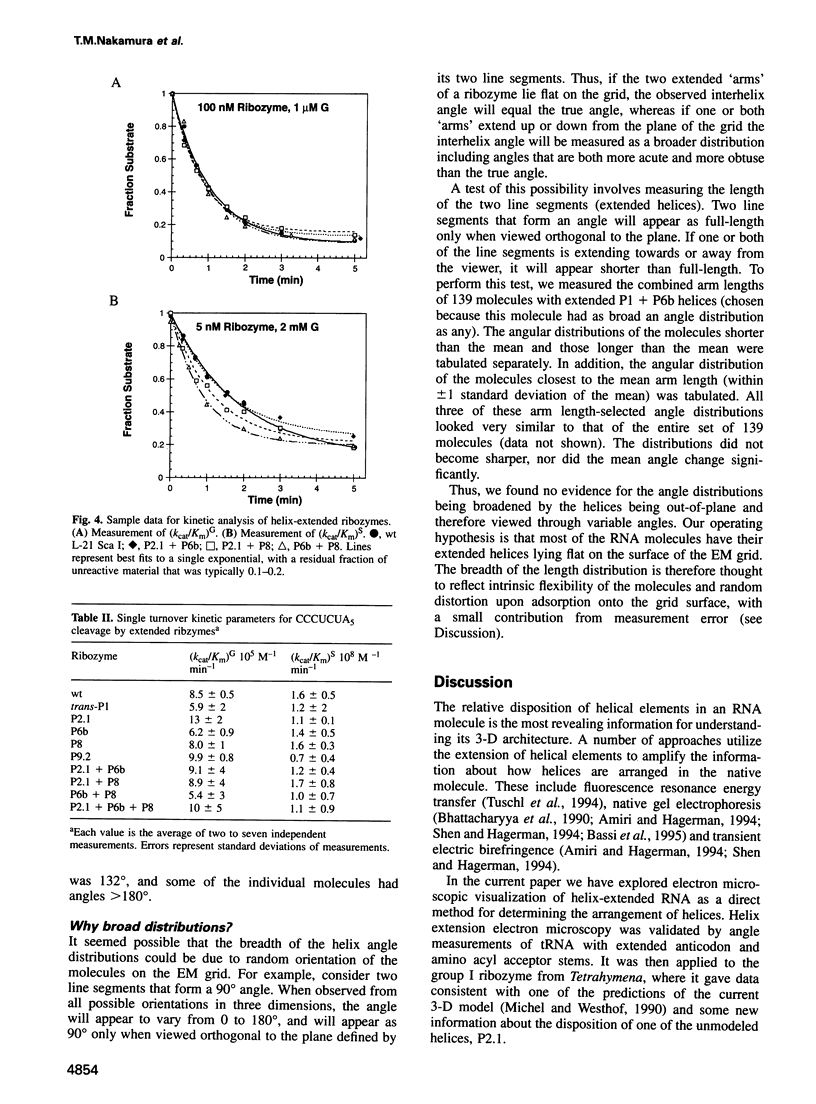
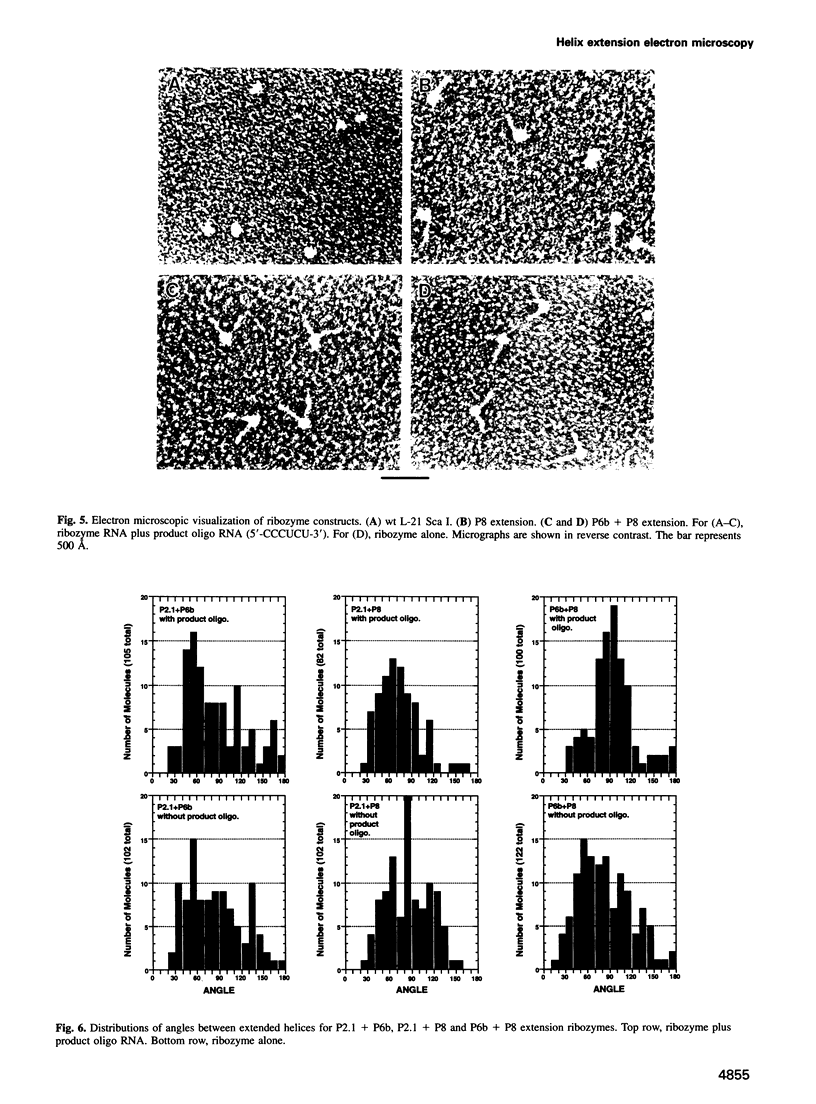
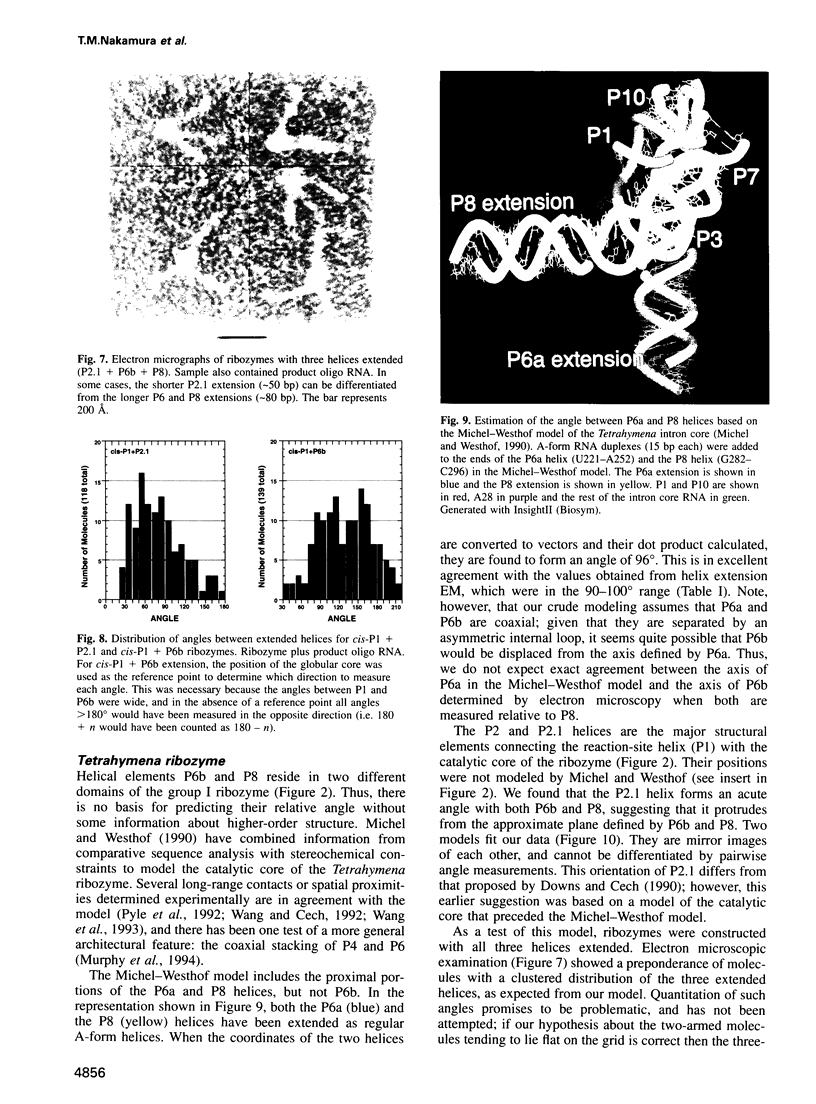
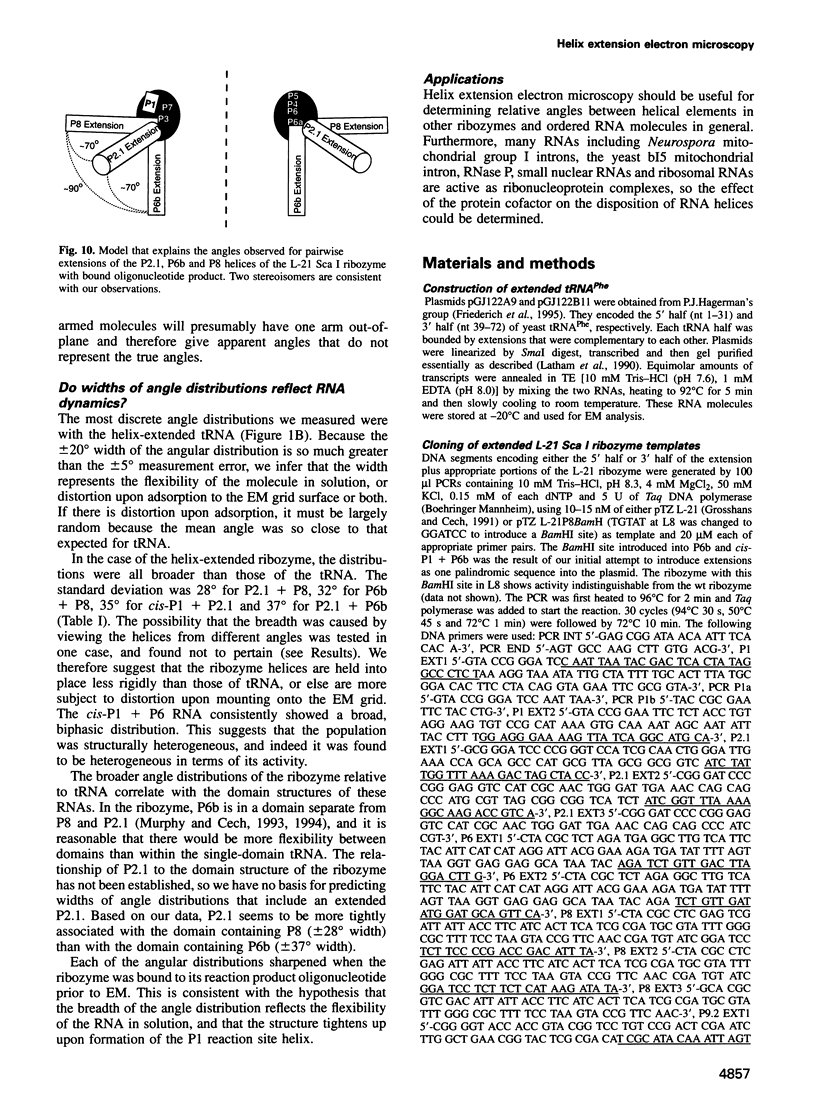
The relative orientation of helical elements in a folded RNA molecule provides key information about its three-dimensional architecture. We have developed a method that involves extending peripheral helices of an RNA, mounting for electron microscopy in the absence of protein and measuring interhelical angles. As a control, extended anticodon and acceptor stems of tRNA(Phe) were found to form a 92 +/- 20 degrees angle, consistent with the X-ray structure. Single, double and triple extensions (50-80 bp) of helical elements P2.1, P6b and P8 of the Tetrahymena group I ribozyme did not alter its catalytic activity. The measured angle between P6b and P8 is consistent with the Michel-Westhof structural model, while the P2.1-P6b and P2.1-P8 angles allow P2.1 to be positioned in the model. The angle distributions of the ribozyme are broader than those of the tRNA, which may reflect the dynamics of the RNA. Helix extension allows low-resolution electron microscopy to provide much higher resolution information about the disposition of helical elements in RNA. It should be applicable to diverse RNAs and ribonucleoprotein complexes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amiri K. M., Hagerman P. J. Global conformation of a self-cleaving hammerhead RNA. Biochemistry. 1994 Nov 15;33(45):13172–13177. doi: 10.1021/bi00249a003. [DOI] [PubMed] [Google Scholar]
- Bassi G. S., Møllegaard N. E., Murchie A. I., von Kitzing E., Lilley D. M. Ionic interactions and the global conformations of the hammerhead ribozyme. Nat Struct Biol. 1995 Jan;2(1):45–55. doi: 10.1038/nsb0195-45. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A., Murchie A. I., Lilley D. M. RNA bulges and the helical periodicity of double-stranded RNA. Nature. 1990 Feb 1;343(6257):484–487. doi: 10.1038/343484a0. [DOI] [PubMed] [Google Scholar]
- Brimacombe R., Atmadja J., Stiege W., Schüler D. A detailed model of the three-dimensional structure of Escherichia coli 16 S ribosomal RNA in situ in the 30 S subunit. J Mol Biol. 1988 Jan 5;199(1):115–136. doi: 10.1016/0022-2836(88)90383-x. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Damberger S. H., Gutell R. R. Representation of the secondary and tertiary structure of group I introns. Nat Struct Biol. 1994 May;1(5):273–280. doi: 10.1038/nsb0594-273. [DOI] [PubMed] [Google Scholar]
- Cech T. R., Herschlag D., Piccirilli J. A., Pyle A. M. RNA catalysis by a group I ribozyme. Developing a model for transition state stabilization. J Biol Chem. 1992 Sep 5;267(25):17479–17482. [PubMed] [Google Scholar]
- Cech T. R. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- Celander D. W., Cech T. R. Visualizing the higher order folding of a catalytic RNA molecule. Science. 1991 Jan 25;251(4992):401–407. doi: 10.1126/science.1989074. [DOI] [PubMed] [Google Scholar]
- Collins J. Instability of palindromic DNA in Escherichia coli. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):409–416. doi: 10.1101/sqb.1981.045.01.055. [DOI] [PubMed] [Google Scholar]
- Damberger S. H., Gutell R. R. A comparative database of group I intron structures. Nucleic Acids Res. 1994 Sep;22(17):3508–3510. doi: 10.1093/nar/22.17.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs W. D., Cech T. R. An ultraviolet-inducible adenosine-adenosine cross-link reflects the catalytic structure of the Tetrahymena ribozyme. Biochemistry. 1990 Jun 12;29(23):5605–5613. doi: 10.1021/bi00475a027. [DOI] [PubMed] [Google Scholar]
- Friederich M. W., Gast F. U., Vacano E., Hagerman P. J. Determination of the angle between the anticodon and aminoacyl acceptor stems of yeast phenylalanyl tRNA in solution. Proc Natl Acad Sci U S A. 1995 May 23;92(11):4803–4807. doi: 10.1073/pnas.92.11.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans C. A., Cech T. R. A hammerhead ribozyme allows synthesis of a new form of the Tetrahymena ribozyme homogeneous in length with a 3' end blocked for transesterification. Nucleic Acids Res. 1991 Jul 25;19(14):3875–3880. doi: 10.1093/nar/19.14.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M. E., Nolan J. M., Malhotra A., Brown J. W., Harvey S. C., Pace N. R. Use of photoaffinity crosslinking and molecular modeling to analyze the global architecture of ribonuclease P RNA. EMBO J. 1994 Sep 1;13(17):3953–3963. doi: 10.1002/j.1460-2075.1994.tb06711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D., Cech T. R. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990 Nov 6;29(44):10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Suddath F. L., Quigley G. J., McPherson A., Sussman J. L., Wang A. H., Seeman N. C., Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974 Aug 2;185(4149):435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Latham J. A., Cech T. R. Defining the inside and outside of a catalytic RNA molecule. Science. 1989 Jul 21;245(4915):276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- Latham J. A., Zaug A. J., Cech T. R. Self-splicing and enzymatic cleavage of RNA by a group I intervening sequence. Methods Enzymol. 1990;181:558–569. doi: 10.1016/0076-6879(90)81151-j. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. In vivo consequences of plasmid topology. Nature. 1981 Jul 23;292(5821):380–382. doi: 10.1038/292380a0. [DOI] [PubMed] [Google Scholar]
- Malhotra A., Harvey S. C. A quantitative model of the Escherichia coli 16 S RNA in the 30 S ribosomal subunit. J Mol Biol. 1994 Jul 22;240(4):308–340. doi: 10.1006/jmbi.1994.1448. [DOI] [PubMed] [Google Scholar]
- McConnell T. S., Cech T. R., Herschlag D. Guanosine binding to the Tetrahymena ribozyme: thermodynamic coupling with oligonucleotide binding. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8362–8366. doi: 10.1073/pnas.90.18.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989 May 19;57(4):585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- Murphy F. L., Cech T. R. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry. 1993 May 25;32(20):5291–5300. doi: 10.1021/bi00071a003. [DOI] [PubMed] [Google Scholar]
- Murphy F. L., Cech T. R. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J Mol Biol. 1994 Feb 11;236(1):49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- Murphy F. L., Wang Y. H., Griffith J. D., Cech T. R. Coaxially stacked RNA helices in the catalytic center of the Tetrahymena ribozyme. Science. 1994 Sep 16;265(5179):1709–1712. doi: 10.1126/science.8085157. [DOI] [PubMed] [Google Scholar]
- Pley H. W., Flaherty K. M., McKay D. B. Three-dimensional structure of a hammerhead ribozyme. Nature. 1994 Nov 3;372(6501):68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- Price J. V., Kieft G. L., Kent J. R., Sievers E. L., Cech T. R. Sequence requirements for self-splicing of the Tetrahymena thermophila pre-ribosomal RNA. Nucleic Acids Res. 1985 Mar 25;13(6):1871–1889. doi: 10.1093/nar/13.6.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle A. M., Murphy F. L., Cech T. R. RNA substrate binding site in the catalytic core of the Tetrahymena ribozyme. Nature. 1992 Jul 9;358(6382):123–128. doi: 10.1038/358123a0. [DOI] [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974 Aug 16;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Shen Z., Hagerman P. J. Conformation of the central, three-helix junction of the 5 S ribosomal RNA of Sulfolobus acidocaldarius. J Mol Biol. 1994 Aug 19;241(3):415–430. doi: 10.1006/jmbi.1994.1517. [DOI] [PubMed] [Google Scholar]
- Stern S., Weiser B., Noller H. F. Model for the three-dimensional folding of 16 S ribosomal RNA. J Mol Biol. 1988 Nov 20;204(2):447–481. doi: 10.1016/0022-2836(88)90588-8. [DOI] [PubMed] [Google Scholar]
- Szostak J. W. Enzymatic activity of the conserved core of a group I self-splicing intron. Nature. 1986 Jul 3;322(6074):83–86. doi: 10.1038/322083a0. [DOI] [PubMed] [Google Scholar]
- Tanner N. K., Schaff S., Thill G., Petit-Koskas E., Crain-Denoyelle A. M., Westhof E. A three-dimensional model of hepatitis delta virus ribozyme based on biochemical and mutational analyses. Curr Biol. 1994 Jun 1;4(6):488–498. doi: 10.1016/s0960-9822(00)00109-3. [DOI] [PubMed] [Google Scholar]
- Tuschl T., Gohlke C., Jovin T. M., Westhof E., Eckstein F. A three-dimensional model for the hammerhead ribozyme based on fluorescence measurements. Science. 1994 Nov 4;266(5186):785–789. doi: 10.1126/science.7973630. [DOI] [PubMed] [Google Scholar]
- Wang J. F., Cech T. R. Tertiary structure around the guanosine-binding site of the Tetrahymena ribozyme. Science. 1992 Apr 24;256(5056):526–529. doi: 10.1126/science.1315076. [DOI] [PubMed] [Google Scholar]
- Wang J. F., Downs W. D., Cech T. R. Movement of the guide sequence during RNA catalysis by a group I ribozyme. Science. 1993 Apr 23;260(5107):504–508. doi: 10.1126/science.7682726. [DOI] [PubMed] [Google Scholar]
- Wang Y. H., Murphy F. L., Cech T. R., Griffith J. D. Visualization of a tertiary structural domain of the Tetrahymena group I intron by electron microscopy. J Mol Biol. 1994 Feb 11;236(1):64–71. doi: 10.1006/jmbi.1994.1118. [DOI] [PubMed] [Google Scholar]
- Waring R. B., Scazzocchio C., Brown T. A., Davies R. W. Close relationship between certain nuclear and mitochondrial introns. Implications for the mechanism of RNA splicing. J Mol Biol. 1983 Jul 5;167(3):595–605. doi: 10.1016/s0022-2836(83)80100-4. [DOI] [PubMed] [Google Scholar]
- Westhof E., Altman S. Three-dimensional working model of M1 RNA, the catalytic RNA subunit of ribonuclease P from Escherichia coli. Proc Natl Acad Sci U S A. 1994 May 24;91(11):5133–5137. doi: 10.1073/pnas.91.11.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Romby P., Romaniuk P. J., Ebel J. P., Ehresmann C., Ehresmann B. Computer modeling from solution data of spinach chloroplast and of Xenopus laevis somatic and oocyte 5 S rRNAs. J Mol Biol. 1989 May 20;207(2):417–431. doi: 10.1016/0022-2836(89)90264-7. [DOI] [PubMed] [Google Scholar]
- Zaug A. J., Been M. D., Cech T. R. The Tetrahymena ribozyme acts like an RNA restriction endonuclease. Nature. 1986 Dec 4;324(6096):429–433. doi: 10.1038/324429a0. [DOI] [PubMed] [Google Scholar]