Abstract
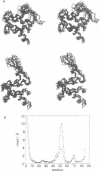
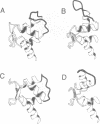
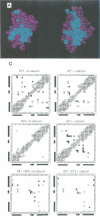
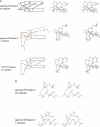
Calcium is a universally employed cytosolic messenger in eukaryotic cells. Most of the proteins that bind signalling calcium are members of the calmodulin superfamily and share two or more helix-loop-helix motifs known as EF-hands. A model, based on structure comparison of different domains and supported by preliminary NMR data, has suggested that EF-hands involved in signal transduction undergo a major conformational change upon calcium binding from a 'closed' to an 'open' state allowing protein-protein interaction. We have determined the solution structures of the EF-hand pair from alpha-spectrin in the absence and in the presence of calcium. The structures are in the closed and open conformation respectively, providing a definite experimental proof for the closed-to-open model. Our results allow formulation of the rules which govern the movement induced by calcium. These rules may be generalized to other EF-hands since the key residues involved are conserved within the calmodulin family.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Babu Y. S., Sack J. S., Greenhough T. J., Bugg C. E., Means A. R., Cook W. J. Three-dimensional structure of calmodulin. Nature. 1985 May 2;315(6014):37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- Bennett V. Spectrin: a structural mediator between diverse plasma membrane proteins and the cytoplasm. Curr Opin Cell Biol. 1990 Feb;2(1):51–56. doi: 10.1016/s0955-0674(05)80030-4. [DOI] [PubMed] [Google Scholar]
- Diamond R. On the multiple simultaneous superposition of molecular structures by rigid body transformations. Protein Sci. 1992 Oct;1(10):1279–1287. doi: 10.1002/pro.5560011006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn B. E., Drakenberg T., Forsén S. The structure of apo-calmodulin. A 1H NMR examination of the carboxy-terminal domain. FEBS Lett. 1993 Dec 27;336(2):368–374. doi: 10.1016/0014-5793(93)80839-m. [DOI] [PubMed] [Google Scholar]
- Gagné S. M., Tsuda S., Li M. X., Chandra M., Smillie L. B., Sykes B. D. Quantification of the calcium-induced secondary structural changes in the regulatory domain of troponin-C. Protein Sci. 1994 Nov;3(11):1961–1974. doi: 10.1002/pro.5560031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güntert P., Braun W., Wüthrich K. Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J Mol Biol. 1991 Feb 5;217(3):517–530. doi: 10.1016/0022-2836(91)90754-t. [DOI] [PubMed] [Google Scholar]
- Güntert P., Wüthrich K. Improved efficiency of protein structure calculations from NMR data using the program DIANA with redundant dihedral angle constraints. J Biomol NMR. 1991 Nov;1(4):447–456. doi: 10.1007/BF02192866. [DOI] [PubMed] [Google Scholar]
- Herzberg O., Moult J., James M. N. A model for the Ca2+-induced conformational transition of troponin C. A trigger for muscle contraction. J Biol Chem. 1986 Feb 25;261(6):2638–2644. [PubMed] [Google Scholar]
- Ikura M., Clore G. M., Gronenborn A. M., Zhu G., Klee C. B., Bax A. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992 May 1;256(5057):632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- Kördel J., Forsén S., Chazin W. J. 1H NMR sequential resonance assignments, secondary structure, and global fold in solution of the major (trans-Pro43) form of bovine calbindin D9k. Biochemistry. 1989 Aug 22;28(17):7065–7074. doi: 10.1021/bi00443a043. [DOI] [PubMed] [Google Scholar]
- Luna E. J. Molecular links between the cytoskeleton and membranes. Curr Opin Cell Biol. 1991 Feb;3(1):120–126. doi: 10.1016/0955-0674(91)90174-w. [DOI] [PubMed] [Google Scholar]
- Meador W. E., Means A. R., Quiocho F. A. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science. 1992 Aug 28;257(5074):1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- Piotto M., Saudek V., Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992 Nov;2(6):661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- Skelton N. J., Forsén S., Chazin W. J. 1H NMR resonance assignments, secondary structure, and global fold of Apo bovine calbindin D9k. Biochemistry. 1990 Jun 19;29(24):5752–5761. doi: 10.1021/bi00476a016. [DOI] [PubMed] [Google Scholar]
- Skelton N. J., Kördel J., Akke M., Forsén S., Chazin W. J. Signal transduction versus buffering activity in Ca(2+)-binding proteins. Nat Struct Biol. 1994 Apr;1(4):239–245. doi: 10.1038/nsb0494-239. [DOI] [PubMed] [Google Scholar]
- Strynadka N. C., James M. N. Crystal structures of the helix-loop-helix calcium-binding proteins. Annu Rev Biochem. 1989;58:951–998. doi: 10.1146/annurev.bi.58.070189.004511. [DOI] [PubMed] [Google Scholar]
- Sundaralingam M., Bergstrom R., Strasburg G., Rao S. T., Roychowdhury P., Greaser M., Wang B. C. Molecular structure of troponin C from chicken skeletal muscle at 3-angstrom resolution. Science. 1985 Feb 22;227(4689):945–948. doi: 10.1126/science.3969570. [DOI] [PubMed] [Google Scholar]
- Taylor K. A., Taylor D. W. Projection image of smooth muscle alpha-actinin from two-dimensional crystals formed on positively charged lipid layers. J Mol Biol. 1993 Mar 5;230(1):196–205. doi: 10.1006/jmbi.1993.1136. [DOI] [PubMed] [Google Scholar]
- Travé G., Pastore A., Hyvönen M., Saraste M. The C-terminal domain of alpha-spectrin is structurally related to calmodulin. Eur J Biochem. 1995 Jan 15;227(1-2):35–42. doi: 10.1111/j.1432-1033.1995.tb20357.x. [DOI] [PubMed] [Google Scholar]
- Viel A., Branton D. Interchain binding at the tail end of the Drosophila spectrin molecule. Proc Natl Acad Sci U S A. 1994 Nov 8;91(23):10839–10843. doi: 10.1073/pnas.91.23.10839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990 Mar;8(1):52-6, 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- Wasenius V. M., Saraste M., Salvén P., Erämaa M., Holm L., Lehto V. P. Primary structure of the brain alpha-spectrin. J Cell Biol. 1989 Jan;108(1):79–93. doi: 10.1083/jcb.108.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S., Sykes B. D., Richards F. M. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol. 1991 Nov 20;222(2):311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- Wishart D. S., Sykes B. D., Richards F. M. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992 Feb 18;31(6):1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- Witke W., Hofmann A., Köppel B., Schleicher M., Noegel A. A. The Ca(2+)-binding domains in non-muscle type alpha-actinin: biochemical and genetic analysis. J Cell Biol. 1993 May;121(3):599–606. doi: 10.1083/jcb.121.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]