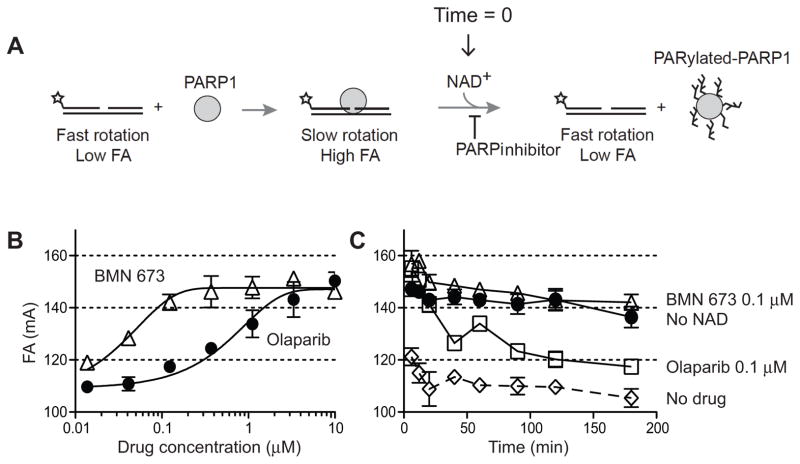
Figure 5. Biochemical trapping of PARP1 by BMN 673.
(A) Scheme of the fluorescence anisotropy (FA) binding assay. The star indicates the site labeled on the DNA substrate with Alexa Fluor488. Unbound nicked DNA substrate rotates fast and gives low FA. PARP1 binding to the substrate slows the rotation and gives high FA. Addition of NAD+ leads to PARP1 dissociation from DNA due to autoPARylation. (B) Concentration-dependent PARP1-DNA association in the presence of BMN 673 or olaparib. FA was measured 40 min after adding NAD+. (C) Time-course of PARP1-DNA dissociation in the presence of BMN 673 and olaparib (0.12 μM each). Addition of NAD+ in the absence of PARP inhibitor immediately reduces PARP1-DNA complexes (DMSO control). In the absence of NAD+, PARP1-DNA complexes remain stable for at least 120 min (no NAD+).