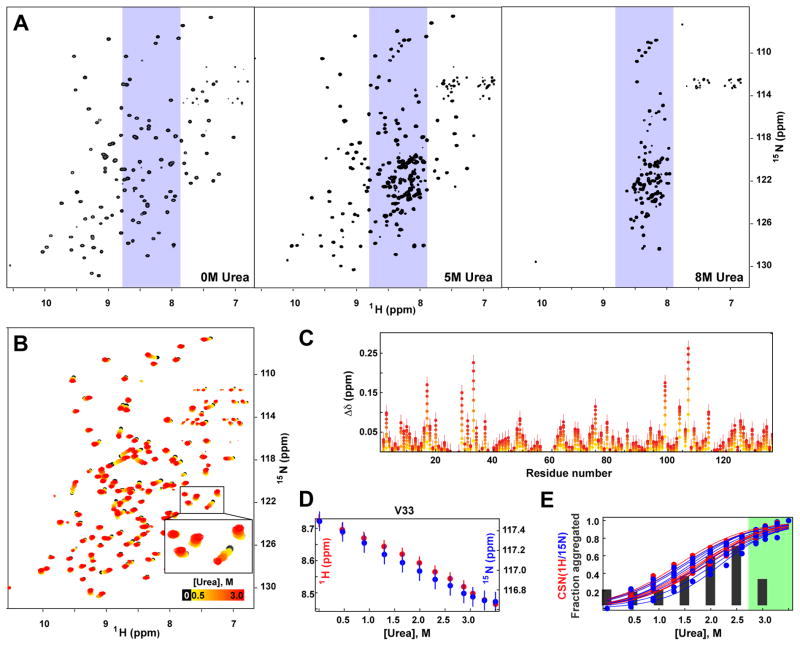
Figure 3.
Urea results in two transitions in CRABP1 WT*: near-native transition at low urea concentrations and protein unfolding at high urea concentrations. A. 1H,15N HSQC NMR spectra of CRABP1 WT* in the absence (left) and presence of 5 M (middle) and 8 M (right) of urea. B. 1H,15N HSQC NMR spectra of CRABP1 WT* in the absence (black) and presence of 0.5–3.1 M urea (colored from yellow to red as the urea concentration increased). C. Amide proton chemical shift (CS) perturbations upon the presence of 0.5–3.1 M urea, mod[Δδ(urea) − Δδ (no urea)]; colored as in B. D. CS dependences of amide 1H (red) and 15N (blue) as a function of urea concentration for a representative residue (V33); see also Fig. S1. E. Aggregation propensity of CRABP1 WT* (gray bars, same as red bars in Figure 2) and fractions of the aggregation-prone near-native conformation (calculated as normalized CS dependences of amide 1H (red) and 15N (blue) [Δδ(urea) − Δδ(no urea)]/[Δδ(3.5 M urea) − Δδ(no urea)]; individual data points correspond to particular highly affected residues), as a function of urea concentration; see also Fig. S1. A green region indicates the higher urea concentrations that result in aggregate instability.