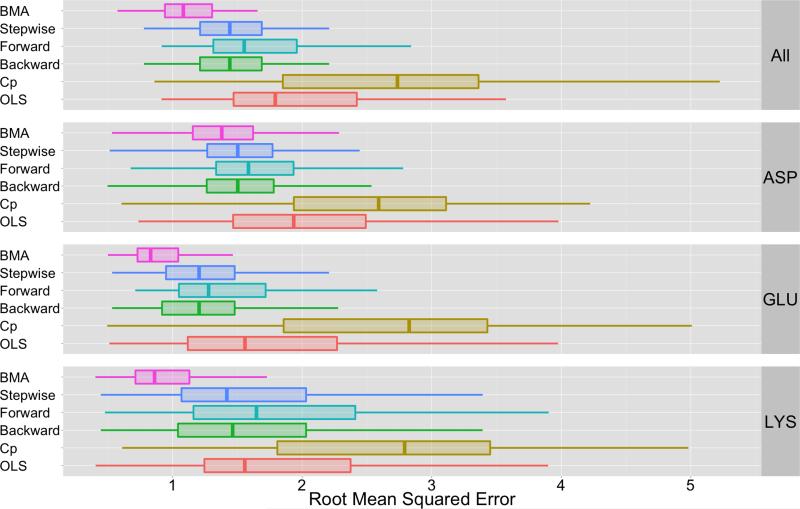
Figure 3.
This figure depicts a summary of root-mean-squared error for different ensemble approaches. The label “BMA” indicates that the BMA aggregate uses all methods from Table II to construct βBMA. The other approaches are alternative ensemble techniques that are based on different strategies for model specification (see Section 3.3). The mean RMSE values shown in this figure are listed in Table V and the statistical significance of BMA’s performance is based on p-values shown in Table VIII. Based on data in Table VIII and Table V, BMA-based approach outperform all other ensemble techniques: BMA-based estimates reduce error by approximately 27% in comparison to Backward and Stepwise regression techniques and by 35% in comparison to Forward regression. In comparison to an ordinary least squares approach that uses all methods in II, BMA reduces error by approximately 46%. Finally, in comparison to Cp methods, BMA reduces error by approximately 60%.