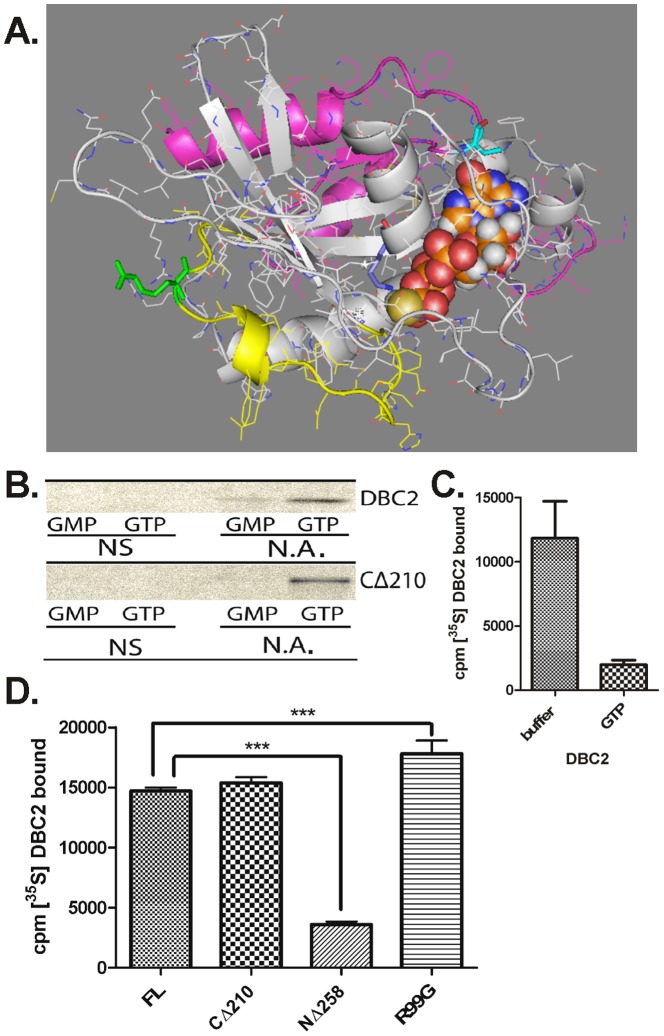
Figure 3. GTP binding activity of DBC2 and its Rho domain.
(A) Homology model of the DBC2 Rho domain was built using Swiss-model and the crystal structure of Rnd1 (2CLS) as a template. The 46 residues which are deleted when DBC2's Rho-domain is truncated to a Ras domain including the loss of the G5 loop [32] are colored in magenta. The switch II region is colored yellow with the R99 residue in green. GTPgammaS is shown as spheres with V191 colored in cyan and K27 in light blue. The image was rendered by PyMol, Delano scientific. (B) [35S]- Flag-tagged DBC2 and CΔ210 were synthesized by TnT in the absence of any additions (no additions, N.A.). TnT RRLs containing empty vector were used as controls for non-specific binding (NS). [35S]DBC2 was pulled down from RRL with GMP or GTP-linked agarose, washed with P100T and analyzed by SDS-PAGE, and autoradiography, as described under Materials and methods. (C) [35S]-Flag-tagged DBC2 was synthesized by TnT in the absence of any additions. The samples were diluted with 5 volumes of TBS (buffer) or TBS containing GTP (GTP) to give a final concentration of 20 mM GTP. After 45 min of incubation with GTP-agarose at 4°C, the samples were washed and the amount of [35S]-DBC2 was determined by scintillation counting. The experiment was carried out in triplicate. (D) [35S]-Labeled wild-type DBC2, DBC2 Rho domain (CΔ210), DBC2ΔRho (NΔ258), and the DBC2/R99G (R99G) mutant were synthesized by TnT, pulled down from RRL with GTP-linked agarose, and washed with P100T as described under Materials and methods. The amount of bound [35S]-DBC2 constructs were quantified by scintillation counting and normalized for the number of Met present in each construct, and for [35S]-Met non-specifically bound to the GTP-agarose. The data represent three independent bio-replicates including three technical replicates. The (***) denotes a significant difference based on a 95% confidence interval, P<0.05.