Figure 4. The effect of Hsp90 inhibitors on the binding of DBC2 to GTP.
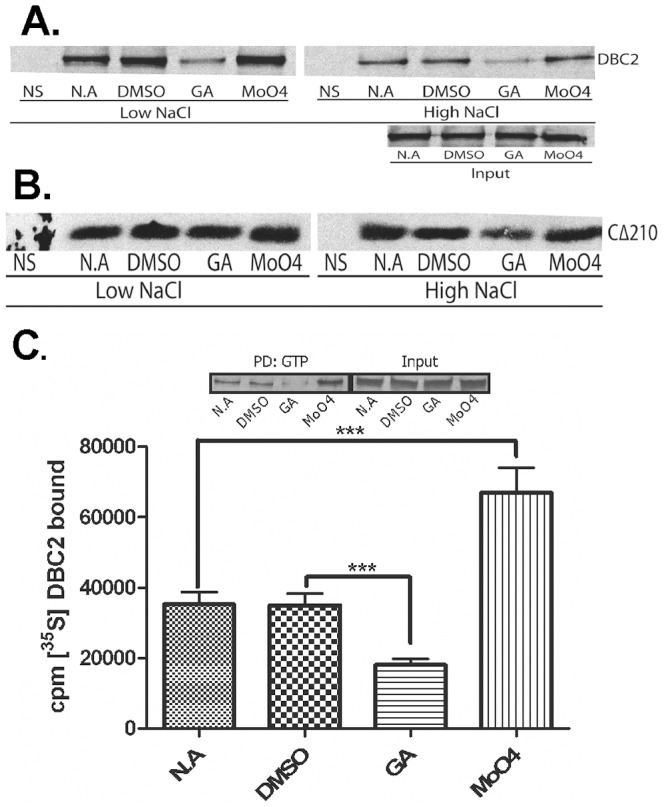
(A) [35S]-Labeled Flag-tagged DBC2 was synthesized in the presence geldanamycin (GA), molybdate (MoO4), or the vehicle controls water (NA) or DMSO, pulled down from RRL with GTP-agarose, washed with buffer containing low (100 mM) or high (500 mM) NaCl, and analyzed by SDS-PAGE and autoradiography, as described under Materials and methods. Naïve RRL containing no template DNA was used as the control for nonspecific binding (A and B: NS). Input: control for amounts of protein that was present in samples used for GTP-agarose pull downs. (B) GTP-agarose pull-downs of [35S]-labeled Flag-tagged DBC2 Rho domain (CΔ210) was carried out in the presence of geldanamycin, molybdate, or vehicle controls as described above in (A) for full length DBC2. (C) GTP-agarose pull downs of [35S]-labeled Flag-tagged DBC2 was carried out in the presence of geldanamycin, molybdate or vehicle controls as described in (A) above. The amount of bound [35S]-DBC2 constructs were quantified by scintillation counting and normalized for [35S]-Met non-specifically bound to the GTP-agarose. The data represent three independent bio-replicates including three technical replicates. The (***) denotes a significant difference based on a 95% confidence interval, P<0.05.
