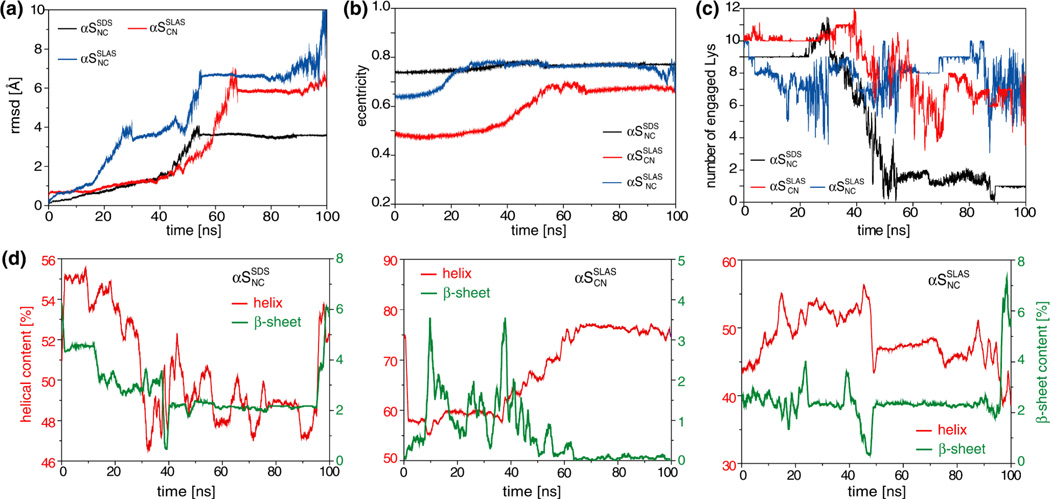
Figure 7.
Evolution of structural parameter during all-atom MD simulations of micelle-bound αS. (a) Root-mean-square-deviation (rmsd) between C[α] coordinates of αS residues 2–92 relative to the starting structures. (b) Micelle eccentricities. (c) Number of lysine ε-NH3+ groups of αS within hydrogen bonding distance (≤ 3.5 Å) to SDS(SO4−) or SLAS(COO−). (d) Contents of helical (α-helix and 310-helix) and β-sheet (β-bridge and β-bulges) secondary structures. Secondary structure was classified with the program DSSP.71