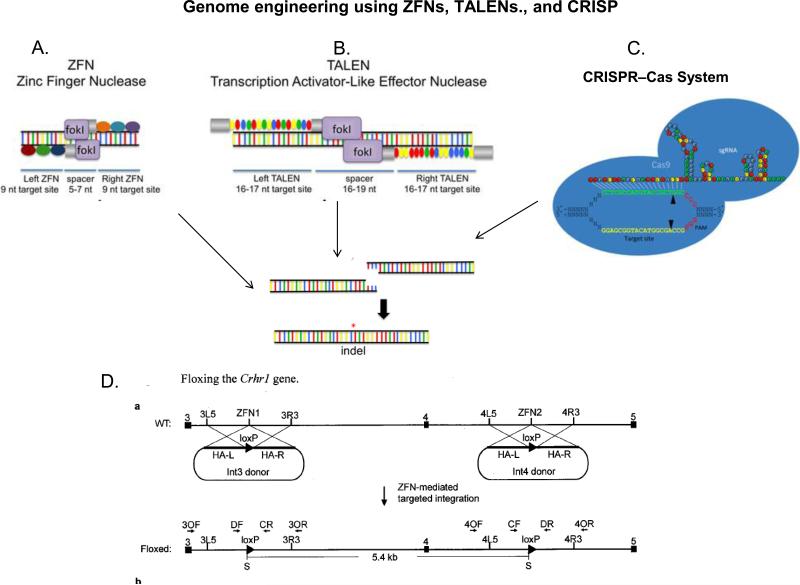
Fig 1. Creating CRE/LoxP rat using Zinc Finger Nuclease, TALEN, and CRISPR technologies for inducible gene knock-in or knock-out.
A. Zinc Finger Nuclease (ZFN) links engineered DNA binding zinc finger (ZF) domain with a DNA-cutting nuclease domain, which contains a Fok1 restriction enzyme site. Each zinc finger recognizes and binds to three targeted nucleotides. The pairing of left ZFN and right ZFN acts like a pair of genomic scissors to produce a double-strand DNA incision in the spacer region. The ZF domains are often extended, doubled or tripled, for longer sequence recognition and increased specificity on each side. ZFNs induced chromosomal breaks are then randomly reconnected by endogenous cellular DNA repair mechanisms called non-homologous end joining, leading to gene knockout. Nonetheless, if there are manipulated DNA strands ending with nucleotides on each side that can form homologous bindings to the DNA break at the time of ZFN incision, a homology dependent repair can occur, resulting in gene knock-in. Adapted with permission151
B. TALENs bind DNA using TAL effector repeat domains derived from Xanthomonas that recognize individual nucleotides. These TALE repeats are ligated together to create binding arrays that recognize extended DNA sequences. The coupled nuclease domain of TALEN cleaves the DNA in the same fashion as ZFN within the intervening spacer region. Adapted with permission151
C. CRISPRs (clusters of regularly interspaced short palindromic repeats) were first found in the Escherichia coli genome likely involved in its immune defense against foreign DNA. CRISPR loci are surrounded by a cohort of conserved CRISPR-associated genes (Cas genes) adjacent to the cluster repeats. The processed CRISPR RNA (crRNAs) serve as sequence-specific guides for the Cas proteins/nucleases to target and generating double strand break (DSB) in the DNAs. In CRISPR genome editing, the crRNA and the trans-activating crRNA (tracrRNA) forms a double strand RNA structure that directs the Cas9 to generate DSBs in the target DNA. At the genomic site where is complementary to the crRNA, the Cas9 HNH nuclease domain cleaves the complementary strand, whereas the Cas9 RuvC-like domain cleaves the non-complementary strand. Reprinted with permission from152.
D. Floxing the Crhr1 gene. In conditional knockin rats, a plasmid DNA that encodes the CRE or LoxP proteins is used together with the genomic editing methods. The plasmid DNA contains homologous binding sequences to bind to the DSB. Insertion of the plasmid DNA is achieved through a mechanism called homologous end joining during DNA repair. This allows CRE-LoxP to be inserted at desired genomic sites to created inducible mutations and generate CRE/LoxP rat for specific neural circuitries in the brain. Figure Courtesy of Dr. Xiaoxia Cui, Sage Labs, St. Louis MO..