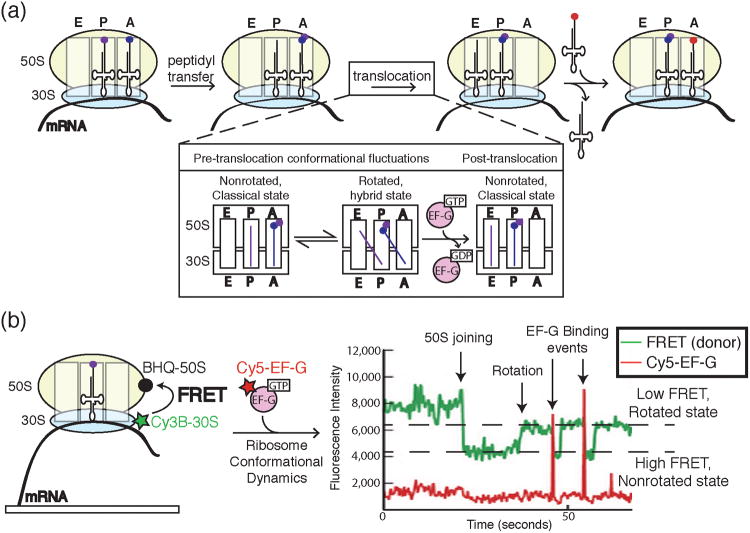
Figure 11.
a) A cartoon representation of key steps in peptide bond formation by the ribosome and translocation along mRNA. The ribosome contains three sites for tRNA binding: the A, P, and E sites. Following peptidyl transfer, the ribosomal subunits undergo conformational dynamics between rotated and non-rotated states. EF-G binding and hydrolysis of GTP leads to successful translocation of the ribosome along the mRNA and concomitant transfer of the P and A site tRNAs to the E and P sites, respectively. The aminoacylated tRNA cognate to the next codon is now free to enter the empty A site. b) Chen et al. correlated ribosome conformation and Cy5-labeled EF-G binding by combining FRET with colocalization assays in ZMWs. Ribosomes were labeled with a FRET donor (Cy3B) in the 30S subunit and FRET quencher (BHQ) in the 50S subunit. Close proximity of Cy3B and BHQ in the nonrotated state caused a decrease in Cy3B signal and an increase in FRET to the quencher. Rotation of the ribosome resulted in decreased FRET and an increase in Cy3B signal. Single molecule data showed ribosomes fluctuating between rotated and nonrotated states with EF-G preferentially binding the rotated state. Images and data were reproduced in this figure from reference 45 and with permission from Nature Publishing Group.