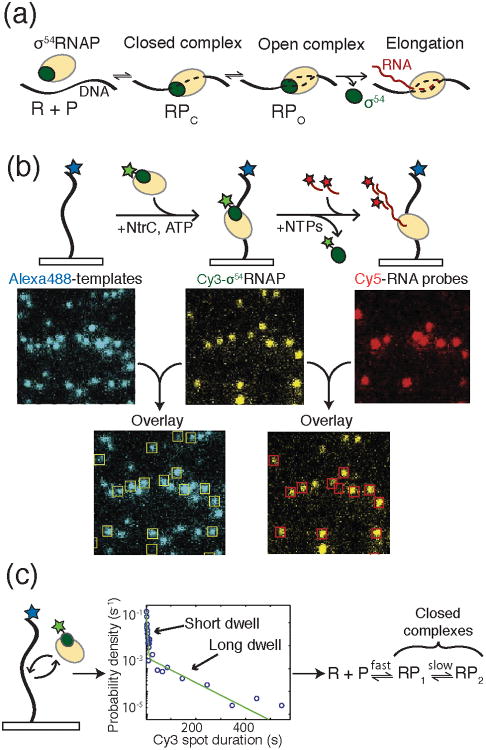
Figure 3.
a) During bacterial transcription initiation, the RNAP holoenzyme first recognizes and binds to promoter DNA to form the closed complex (RPc). Upon DNA melting, open complex (RPo) is formed. Finally, RNAP escapes the promoter to form the elongation complex. b) Experimental setup for CoSMoS analysis of bacterial transcription initiation by Friedman and Gelles. DNA templates were immobilized on a glass surface and labeled with Alexa488™ (blue star). Binding of the σ54-RNAP holoenzyme to the template was monitored by colocalization of the DNA and Cy3-labeled σ54 (green star). Production of RNA was then detected by hybridization of Cy5-labeled DNA oligos and colocalization with the DNA templates and RNAP holoenzyme. c) In an experiment colocalizing RNAP holoenzyme with template DNA in the absence of the NtrC activator, two dwell times were detected following histogram analysis of σ54-RNAP spot duration. The data were consistent with the formation of two closed complexes being present in the initiation pathway. Images and data were reproduced in this figure from reference 13 and with permission from Elsevier.