Figure 2.
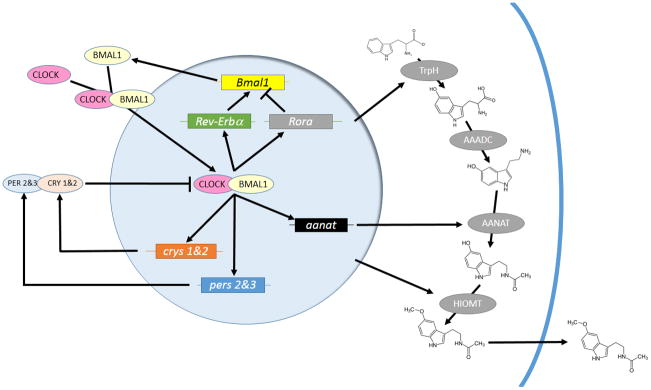
Schematic of the molecular clockworks regulating circadian patterns of melatonin biosynthesis in a pinealocytes or retinal photoreceptor. Positive elements CLOCK and BMAL1 enter the nucleus and activate expression of genes whose promoters contain an E-Box. Among these are the negative elements period 2 & 3 (pers 2&3) and cryptochromes 1& 2 (crys 1&2), Rev-Erbα and Rora, which form a secondary loop regulating Bmal1 transcription, and output, clock-controlled genes such as arylalkylamine-N-actyltransferase (aanat). The pers and crys are translated, form heterodimers with other components, such as the casein kinases, and reenter the nucleus to interfere with CLOCK/BMAL1 activation. Melatonin biosynthesis pathways are indicated on the right. Amino acid tryptophan is converted to 5-hydroxytryptophan by tryptophan hydroxylase (TrpH). Aromatic amino acid decarboxylase (AAADC) then converts 5-hydroxytryptophan to 5-hydroxytryptamine (5HT; serotonin), Then, during the night, AANAT converts 5HT to N-acetylserotonin, a substrate for hydroxyindole-O-methyltransferase (HIOMT), which produces melatonin itself. Presumably, melatonin diffuses out of the cell at this time, although a release mechanism may exist.
