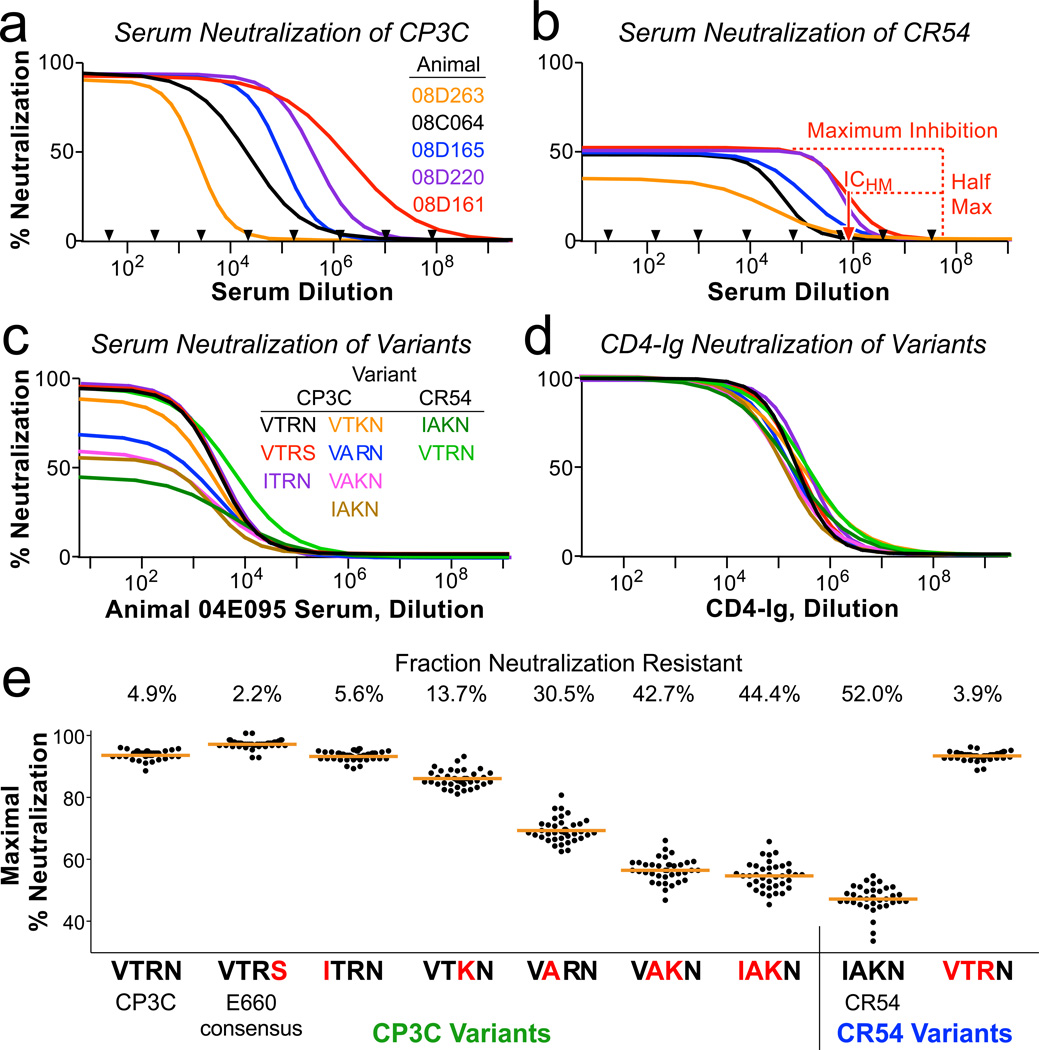
Figure 3. Sequences accounting for neutralization resistance.
a,b, Neutralization curves of CP3C, a sensitive clone from E660 (a), and of CR54, a resistant clone from E660 (b), using dilutions of sera from five Env-immunized animals (selected to show the range of potency). Black arrows indicate which dilutions were tested in duplicate; curves represent non-linear least squares regressions of a four-parameter binding model. Nearly 100% of CP3C virions, but only 40–50% of CR54 virions, can be neutralized by immune sera. Red dashed lines show how ICHM is derived for animal 08D161. c,d, Neutralization curves of 9 viral variants using sera from one animal (c) or CD4-Ig (d). The parent virus into which mutations were made is listed, along with the amino acids at positions 23, 45, 47, and 70. e, All variants were assayed using serial dilutions of sera from all 40 Env-immunized animals. Shown is the maximum fraction of each virus that was neutralized (determined by regression analysis). Blue letters indicate amino acid substitutions compared to the parent virus. The numbers above the graphic indicate the mean resistant fraction for each virus.