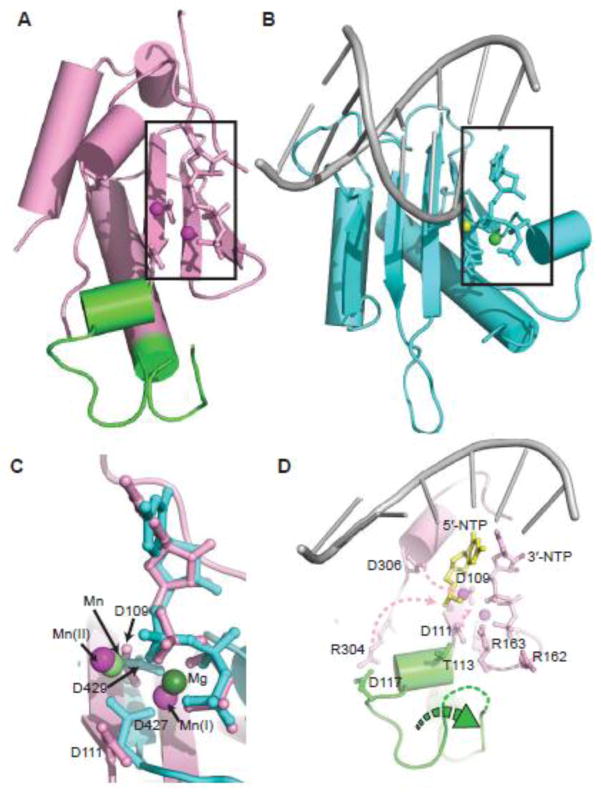
Fig. 5.
Structural alignment of p48 and pol λ catalytic domains and a model for structural changes to activate p48. (A, B) Core catalytic regions from the structures of p48ΔL•UTP•Mn2+ and the pol λ complex, respectively, including conserved catalytic residues, dUpnpp, UTP and primed DNA (PDB ID: 2PFO). Mn2+ ions in (A) are magenta spheres. Mn2+ and Mg2+ ions in (B) are light and dark green spheres, respectively. (C) Overlay of the active sites from (A) and (B). Metals and key acidic residues are labeled. (D) Proposed conformational changes in p48 required to activate primer synthesis, highlighting the zinc-binding motif (green), template ssDNA (gray), 5′-NTP (yellow), and Mn2+ ions (magenta). Pink dashed arrows show the movement of the side chains of catalytically important residues. The green dashed arrow indicates the shift of the zinc-binding motif.