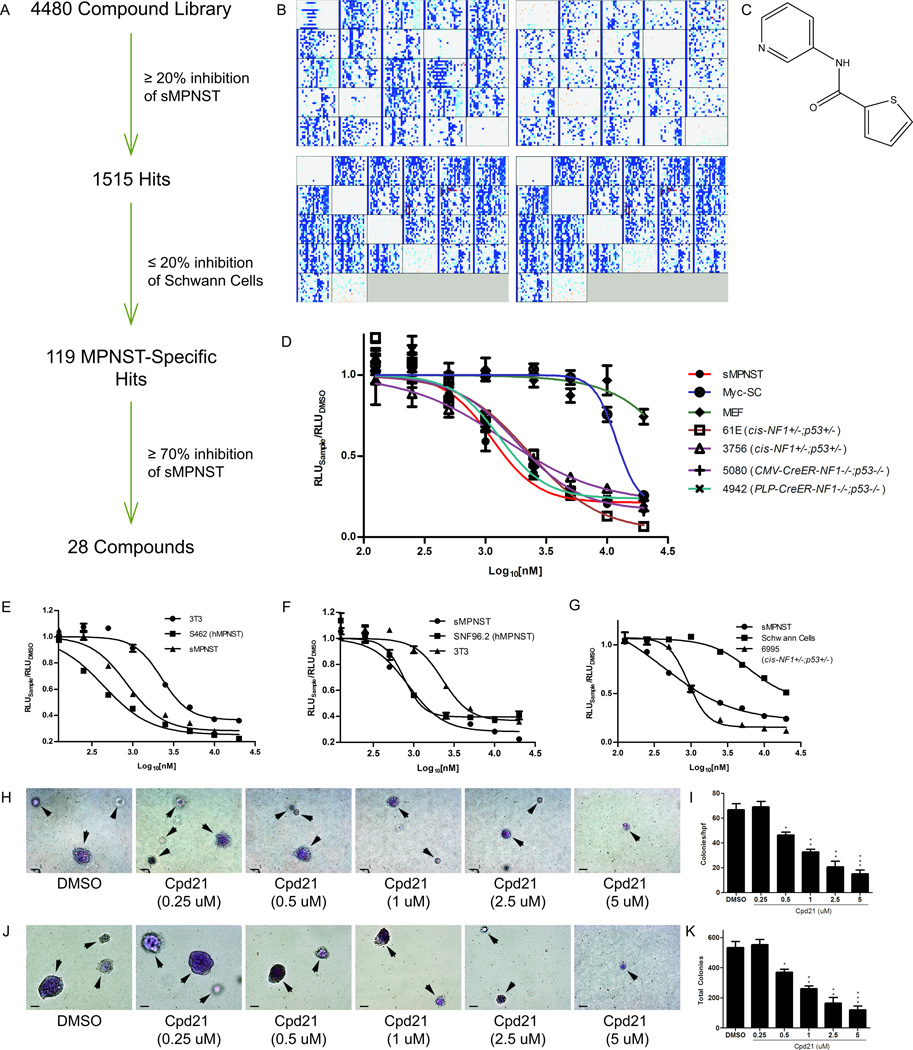
Figure 2. Identification of Cpd21 by high-throughput, small-molecule screening and Validation.
(A) Summary of screening procedure. (B) High-throughput screening data for sMPNST cells (upper panels) and Schwann cells (lower panels). Intensity of blue or red signal correlates with degree of inhibition or induction of ATP levels by small molecules, respectively. (C) Structure of Cpd21. (D) Dose-response curves of Cpd21 (0.125, 0.25, 0.5, 1, 2.5, 5, 10, and 20 µM) in sMPNST cells, MEF, Schwann cells, and MPNST cells from multiple murine models. (E, F) Dose-response curves of Cpd21 treatment on human MPNST cell lines, S462 and SNF96.2, respectively, compared with mouse NIH3T3 cells and sMPNST cells. (G) Dose-response curves of Cpd21 treatment on wildtype Schwann cells, compared with sMPNST and MPNST from cis-Nf1f/f;p53f/f mice. (H) Soft agar assay of Cpd21-treated (0.25, 0.5, 1, 2.5, or 5 µM) sMPNST cells compared with DMSO. Arrows indicate colonies, which may be out of plane in agar. Scale bars represent 100 µm. (I) Quantification of H. (J) Soft agar assay of MPNST cells from cis-Nf1+/−;p53+/− mice, and that were treated with DMSO or Cpd21 at 0.25, 0.5, 1, 2.5, or 5 µM. Arrows indicate colonies, which may be out of plane in agar. Scale bars represent 100 µm. (K) Quantification of J. All values represent the mean±standard deviation. Student’s t-test used for significance testing (*p<0.05, **p<0.01, ***p<0.001).