The various highly compartmentalized, membrane-bound organelles are essential for cellular metabolism in eukaryotic cells.[1,2] The specialized composition of proteins and lipids in different organelles enables them to accomplish specialized processes, and sophisticated sorting mechanisms direct molecules to specific intracellular locations to maintain cellular functions.[2–4] Although it is ubiquitous to life that cells routinely generate and sort nanostructures, including protein complexes, organelles and chromosomes, our ability to similarly engineer and sort synthetic organelles in vivo remains primitive.[2,5,6] To address this challenge, this communication describes an innovative approach for co-assembly and self-sorting of materials inside living cells using genetically-encoded protein polymers – elastin-like polypeptides (ELPs).[7] ELPs are repetitive polypeptides with the sequence of (Val-Pro-Gly-Xaa-Gly)n derived from human tropoelastin, where Xaa and n represent the ‘guest residue’ identity and number of repeat units, respectively.[8,9] ELPs mediate self-assembly by temperature-triggered phase separation above their transition temperature (Tt).[10] Depending on the composition and arrangement of guest residues, monoblock ELPs form micro-structures distinct from diblock ELPs after assembly.[11–13] While monoblock ELPs form large protein coacervates, amphiphilic ELP diblock copolymers sometimes assemble nanoparticles of less than 100 nm in diameter.[14,15] Prior studies revealed that ELP fusion proteins assemble genetically engineered protein microdomains (GEPMs) in living eukaryotic or E. coli cells;[16,17] however, this communication is the first report exploring how ELPs can sort or assemble two distinct proteins. The hypothesis is that different monoblock ELPs with similar transition temperatures may spatially coassemble into mixed GEPMs, and that these will spatially sort from ELP diblock copolymers (Scheme 1). To test this hypothesis, three different monoblock ELPs and one diblock ELP were biosynthesized and purified, and their micro-structures were identified and characterized. The capability of these ELPs to spatially co-assemble and self-sort was evaluated using confocal laser scanning microscopy both in vitro and in the eukaryotic cytosol. These findings reveal a potentially powerful strategy for intracellular co-assembly and sorting of GEPMs, and may have utility in organizing synthetic organelles enriched in distinct functional proteins.
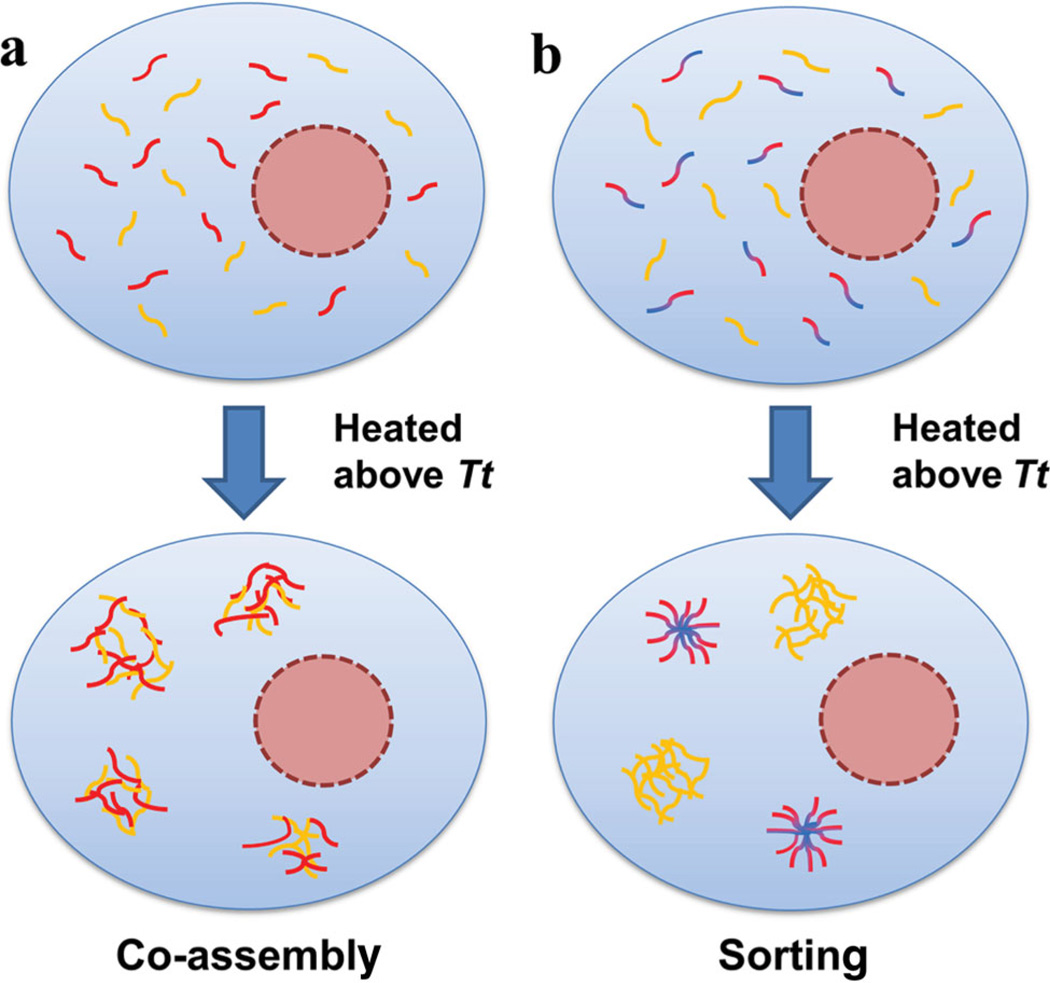
Scheme 1.
Co-assembly versus self-sorting of Genetically Engineered Protein Microdomains (GEPMs) in eukaryotes. a) Two different soluble ELP monoblocks are uniformly distributed in the eukaryotic cytosol when below their Tt. When induced to phase separate, structurally similar ELP monoblocks mix and co-assemble into micron sized mixed GEPMs. b) Structurally distinct ELP monoblock and diblock copolymers are homogeneously mixed below Tt. After transition, the nanoparticles assembled by ELP diblocks self-sort into GEPMs that are spatially separate from the monoblock ELP GEPMs in the eukaryotic cell. This process is tunable, switchable, and reversible; furthermore, because it is based on genetically-encoded protein polymers, it may be useful to drive assembly and sorting of functional proteins in living cells.
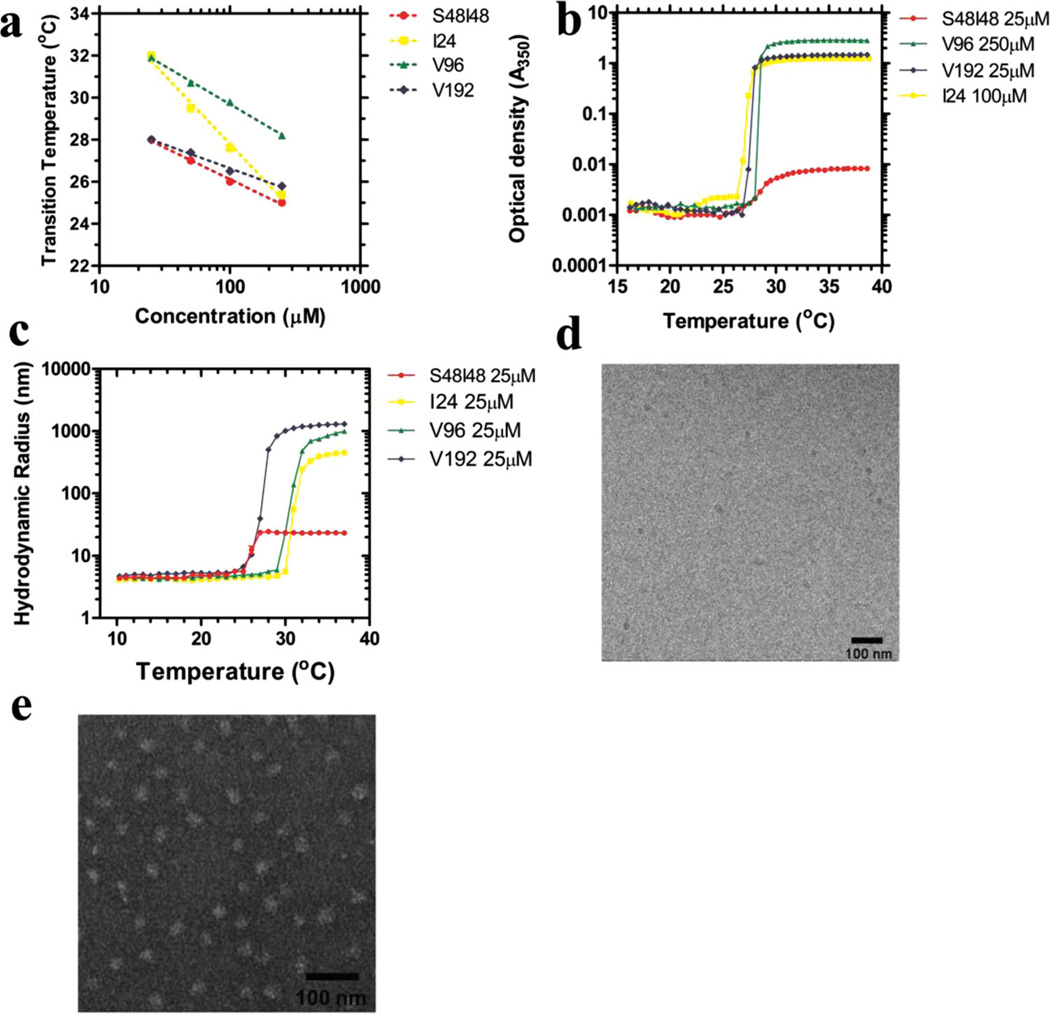
Four ELPs have been biosynthesized and purified (Table 1) in order to mimic the natural protein sorting process in eukaryotes. Among these four ELPs, I24, V96 and V192 are hydrophobic monoblock ELPs with isoleucine or valine as guest residues, while S48I48 is an amphiphilic diblock ELP copolymer with serine and isoleucine as the guest residues of hydrophilic and hydrophobic blocks respectively. The micro-structures formed after their temperature-sensitive transitions were assessed by optical density measurement, dynamic light scattering (DLS), cryogenic-transmission electron microscopy (Cryo-TEM) and negative staining TEM (Figure 1). Optical density measurement using a range of ELP concentrations from 25 µM to 250 µM confirmed a negative correlation between the concentration and transition temperature (Figure 1a). Using this fit, a common transition temperature of about 28 °C was identified for the four ELPs by adjusting the concentration. Another optical density measurement of these four ELPs transitioned at the same temperature (28 °C) with different concentrations suggested the formation of micro-structures with widely differing optical density (Figure 1b). Above the assembly temperature, the high A350 optical densities of I24, V96 and V192 monoblock ELPs were consistent with the formation of large microparticles, while the relatively low optical density of S48I48 was consistent with the formation of nanoparticles. To further explore these micro-structures, DLS, cryo-TEM, and negative staining TEM were performed to measure the hydrodynamic radii and observe morphology of the nanostructures respectively (Figure 1 c–e). DLS analysis determined that amphiphilic S48I48 diblock ELP assembled micelle nanoparticles with a hydrodynamic radius of 24 nm, while the other ELP monoblock coacervates formed large microparticles (hydrodynamic radii from 0.4 to 1.3 µm) above their Tt (Figure 1c). Consistent with DLS data, negative staining TEM imaging also confirmed the assembly of large GEPMs from monoblock ELPs I24, V96 and V192 (Supporting Information, Figure S1). Moreover, the morphology of S48I48 micelle nanoparticles was confirmed by cryo-TEM (Figure 1d) and negative staining TEM (Figure 1e) imaging with an average measured radius of 14.3 ± 1.4 nm and 17.1 ± 1.4 nm, respectively.
Table 1.
ELP protein polymers of different molecular weight, hydrophobicity, and polymer architecture with similar assembly temperatures.
| ELP Nomenclature | Amino acid sequencea) |
Ttb) [°C] |
Molecular Weightc) [Da] |
Morphology above Ttd) | Particle radiuse) [nm] |
|---|---|---|---|---|---|
| S48I48 | G(VPGSG)48 (VPGIG)48Y |
28.0 | 39,644 | nanoparticle | 23.7 ± 0.3 |
| I24 | G(VPGIG)24Y | 32.0 | 10,596 | microparticle | 437.2 ± 15.5 |
| V96 | G(VPGVG)96Y | 31.9 | 40,992 | microparticle | 918.8 ± 76.2 |
| V192 | G(VPGVG)192Y | 28.0 | 81,984 | microparticle | 1291.4 ± 12.7 |
ELP gene sequences were confirmed by DNA sequencing and diagnostic digestions;
Transition temperature by optical density (25 µM, pH 7.4) is indicated for I24, V96 and V192, while the critical micelle temperature (CMT) is indicated for S48I48;
Molecular weight was estimated from open reading frame excluding methionine start codon;
Morphology of ELPs after transition was confirmed by dynamic light scattering (DLS), cryo-TEM and negative staining TEM imaging;
Particle radii of ELP samples (25 µM, pH 7.4) were measured by DLS and presented as Mean ± SD (n = 3).
Figure 1.
Tunable assembly of micro-structures from hydrophobic ELP monoblocks and amphiphilic ELP diblock copolymers. Four ELPs are considered in this study with varying molecular weight, ELP guest residue, and polymer architecture (Table 1). a) The concentration-temperature phase diagram for these distinct polymers enables the selection of concentrations that assemble at a common temperature. All four ELPs transition at 28 °C under different concentrations: S48I48 25 µM, V96 250 µM, V192 25 µM, I24 100 µM. b) Above this temperature, optical density measurements show that ELP monoblocks I24, V96 and V192 form highly turbid particle suspensions, while the ELP diblock S48I48 assembles particles with a minimal increase in optical density. Under optimal conditions, all four constructs assemble at nearly identical temperatures. c) DLS analysis demonstrates that S48I48 assembles into nanoparticles with a hydrodynamic radius of 24 nm, while ELPs monoblocks form larger microparticles ranging from 0.4 to 1.3 µm in radius. d) Cryo-TEM imaging confirms the formation of S48I48 into small nanoparticles with a radius of 14.3 ± 1.4 nm (Mean ± SD, n = 10). Bar length = 100 nm. e) Negative staining TEM image of S48I48 nanoparticles. The measured radius is 17.1 ± 1.4 nm (Mean ± SD, n = 10). Bar length = 100 nm.
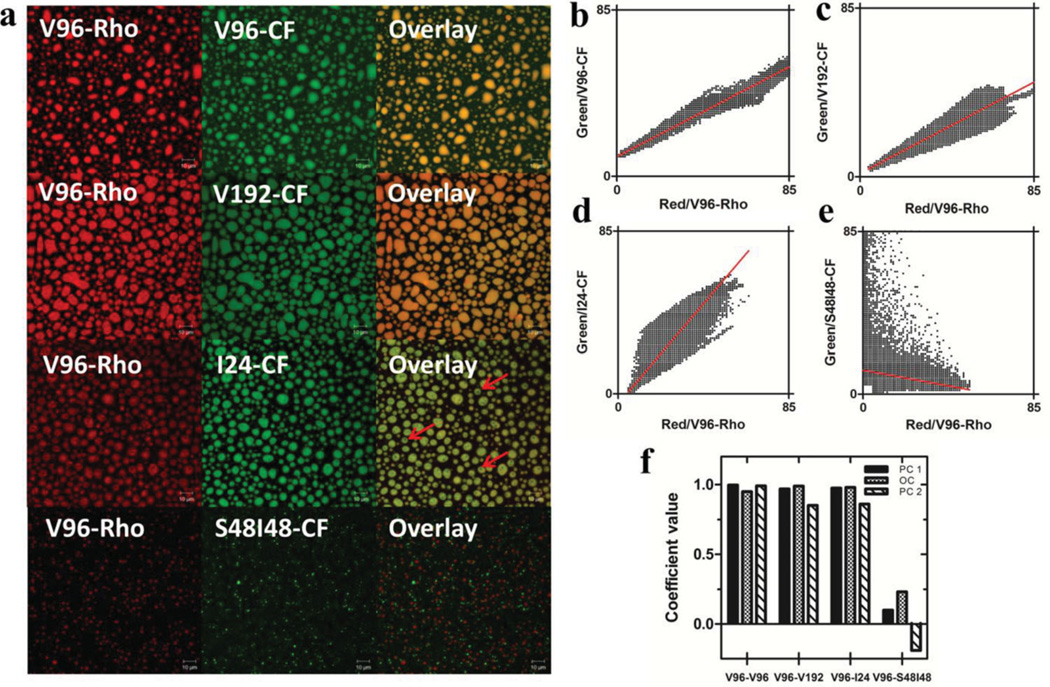
Since distinct micro-structures above Tt have been identified, the capability of monoblock and diblock ELPs to spatially co-assemble and self-sort was first examined in vitro (Figure 2). The assay was developed in a glass bottom dish with a temperature control stage. Using confocal microscopy, ELPs labeled with different fluorophores could be distinguished and analyzed by quantitation of colocalization. Four groups of ELPs were assigned based on their differences in the repetitive unit lengths (n), guest residues (Xaa) and block composition (monoblock vs. diblock). The two ELPs in each group were site-specifically labeled at the amino terminus with rhodamine (Rho) or carboxyfluorescein (CF), mixed in the dish and imaged above their Tt (Figure 2a). Below Tt, each group was uniformly mixed and highly colocalized (Figure S2). Above Tt, large microdomains colocalized in the mixtures of V96-V96, V96-V192 and V96-I24. As expected, mixtures of ELPs with the same guest residue (Xaa = Val) and different molecular weights (n = 96 and 192) yielded nearly optimal spatial co-assembly above their common Tt. Unexpectedly, in the V96-I24 mixture a fraction of I24-CF coacervate forms ‘red free’ domains that are inaccessible to the surrounding V96-Rho coacervate (Figure 2a). This may be due to I24’s higher hydrophobicity (Xaa = Ile vs. Val) and lower molecular weight (1/4 of V96) compared to V96, which may allow it to form a crystalline phase within the amorphous V96 melt. Thus, even though I24 and V96 spatially co-assemble, they form heterogeneous microdomains containing subfeatures. In contrast, the assembled microdomains of V96-Rho are spatially separate from those of S48I48-CF in the V96-S48I48 mixture. Interestingly, in the V96-S48I48 mixture the size of V96 coacervates was significantly smaller than in the other mixtures. Despite their apparent spatial sorting, S48I48 nanoparticles appear to mediate growth of smaller V96 microdomains, which is a phenomenon that will be explored in future studies. Colocalization analysis was performed using software JACoP in ImageJ and LSM510 for all confocal images (Figure 2b–f). Scatter plots displayed dissimilar correlations between red and green pixels in each group (Figure 2b–e). Both positive slopes of the linear regression lines and tight distribution of data points along the lines indicated a positive correlation between the two pixels in the mixtures of V96-V96 (Slope = 0.54, r2 = 0.9939), V96-V192 (Slope = 0.54, r2 = 0.9106) and V96-I24 (Slope = 1.23, r2 = 0.9514) (Figure 2 b–d). However, a negative slope and near-axial distribution of data points was observed in the V96-S48I48 mixture, which may be interpreted as a measure of spatial sorting between these two ELPs (Slope = −0.19, r2 = 0.0361) (Figure 2e). To quantitatively evaluate the degree of colocalization between two ELPs in each group, Pearson's coefficients (PC) were calculated by both JACoP and LSM510, and overlap coefficients (OC) were obtained from JACoP (Figure 2f). A difference in PC between JACoP and LSM510 is noticeable because LSM510 subtracts manually determined background intensity from the image while JACoP does not. The PC for the groups of V96-V96, V96-V192 and V96-I24 were close to 1, which revealed strong colocalization. In contrast, the PC of V96-S48I48 group from both JACoP and LSM510 were close to 0 or even negative, which suggest that these two ELPs self-sort. The OC quantifies the overlap of the two pixels (range from 0 to 1) also suggests strong co-assembly of the three monoblock ELP groups and self-sorting between V96 and S48I48. To further study the relation between self-sorting of monoblock and diblock GEPMs and ELP concentration, different mixing ratios of V96 and S48I48 were tested for in vitro self-sorting. It was discovered that the spatial self-sorting of monoblock and diblock GEPMs was independent of ELP concentration (Figure S3).
Figure 2.
Only monoblock and diblock copolymers spatially sort into distinct GEPMs. a) Confocal microscopy imaging was used to characterize ELP micro-structures above the matched transition temperature. On a glass bottom dish, two purified ELPs labeled respectively with rhodamine (Rho) and carboxyfluorescein (CF) were mixed. Below their transition temperature both colors remain mixed and diffuse (Figure S2). The ‘red free’ domains in the V96-I24 mixture are indicated with red arrows. Scale bar = 10 µm. b–e) Scatter plots of red and green pixels were generated from converted (8-bit) confocal images of ELP mixtures. Linear regression lines are shown in red for each mixture. Noise in each image was determined as Red < 5 and Green < 5 and excluded from the scatter plots. f) For each mixture, Pearson’s coefficients (PC1, PC2) and overlap coefficients (OC) were estimated. PC1 and OC were generated using JACoP software. PC2 was generated from LSM 510 software.
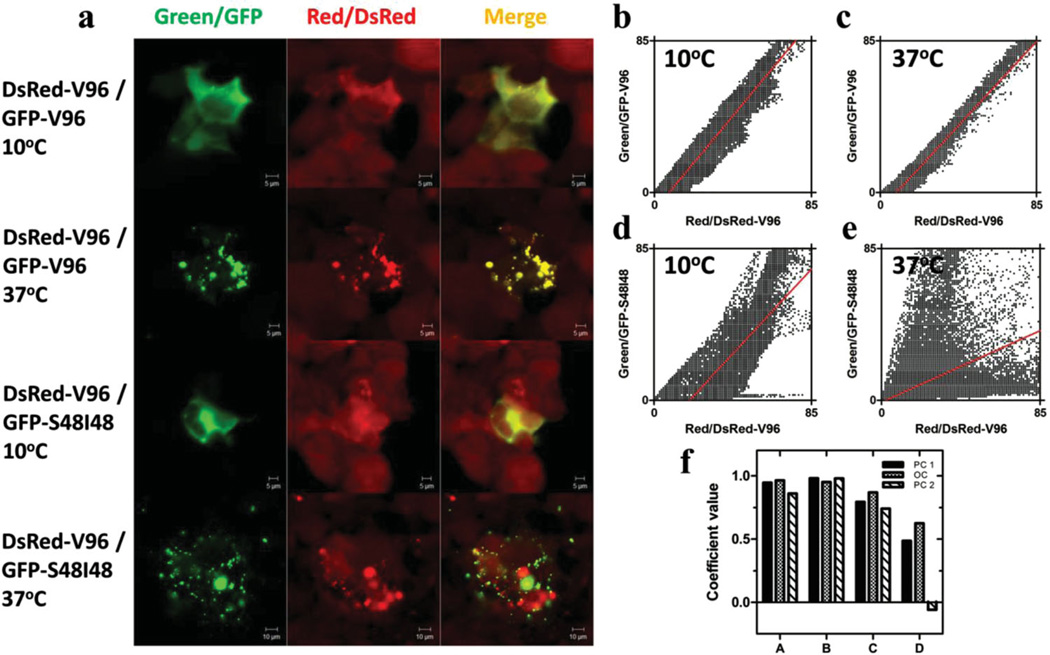
Having obtained evidence of in vitro spatial co-assembly and self-sorting, we hypothesized that the genes encoding for ELP microdomains would have similar behavior when observed in a complex biological environment such as the cytosol of a living eukaryote. A human embryonic kidney (Hek) cell line (Hek-DsRed-V96) was established to stably express DsRed-V96 – a fusion of a red fluorescent protein and monoblock ELP. Mammalian GFP-ELP fusions – GFP-V96 and GFP-S48I48 were cloned and transiently transfected into Hek-DsRed-V96 cells respectively. Confocal microscopy was utilized to image the cells containing both DsRed and GFP fluorescence (Figure 3a). With a temperature control stage, it was determined that all fusion ELPs had a similar intracellular transition temperature (DsRed-V96 26.3 °C, GFP-V96 26.9 °C, and GFP-S48I48 25.6 °C). Therefore, 37 °C and 10 °C were selected to represent intracellular co-assembly and self-sorting after and before microdomain formation. In both groups of DsRed-V96/GFP-V96 and DsRed-V96/GFP-S48I48 at 10 °C, GFP and DsRed fluorescence was uniformly distributed throughout the cell, which was reflected by strong colocalization between the two soluble monomers. ELP microdomains (red and green puncta) assembled in both groups when the temperature was raised to 37 °C. The microdomains of DsRed-V96 and GFP-V96 extensively colocalized with each other while spatially separated microdomains were observed in cells with both DsRed-V96 and GFP-S48I48. Similar to in vitro data analysis, the intracellular confocal images were quantitatively analyzed for pixel colocalization using JACoP and LSM510 software. Consistent with the confocal images, the statistical analysis demonstrated intracellular co-assembly of DsRed-V96 and GFP-V96 and self-sorting between DsRed-V96 and GFP-S48I48 (Figure 3b–f). While remaining soluble at 10 °C, both DsRed-V96/GFP-V96 and DsRed-V96/GFP-S48I48 groups showed positive slopes and high (close to 1) r2 values of the linear regression lines in scatter plots (DsRed-V96/GFP-V96 slope = 1.24, r2 = 0.8958; DsRed-V96/GFP-S48I48 slope = 1.12, r2 = 0.6315) (Figure 3b and d) After phase transition at 37 °C, the value of r2 slightly increased in the group of DsRed-V96/ GFP-V96 (r2 = 0.9642) while a significant decrease was observed in both values of the slope and r2 in DsRed-V96/GFP-S48I48 group (slope = 0.47, r2 = 0.2362) (Figure 3c and e). These data indicated that after phase transition, the DsRed-V96/GFP-V96 group assembled microdomains with a higher degree of colocalization. In contrast, self-sorting DsRed-V96 and GFP-S48I48 microdomains had a much lower the degree of pixel colocalization. Moreover, the high (close to 1) values of PC and OC confirmed co-assembly of the microdomains of DsRed-V96 and GFP-V96. In contrast, the coefficient values dramatically decreased with the formation of DsRed-V96 and GFP-S48I48 microdomains indicating that these two ELP fusion proteins can spatially self-sort in living cells (Figure 3f). In the intracellular assay, PC values from JACoP and LSM510 differ because of the relatively high background intensity resulting from the overexpression of DsRed-V96. Without removing DsRed background, PC from JACoP included colocalization of low intensity background pixels in DsRed-V96 and GFP-S48I48 37 °C group which resulted in a slightly positive value; however, a negative value obtained from LSM510 after removing those unwanted pixels provided stronger quantification of intracellular sorting of DsRed-V96 and GFP-S48I48 microdomains.
Figure 3.
Fluorescent ELP fusion proteins in the eukaryotic cytosol can co-assemble or self-sort GEPMs. a) Confocal microscopy imaging of GFP-ELP and DsRed-ELP fusions in Hek-DsRed-V96 cells. GFP-V96 and GFP-S48I48 were respectively transfected and expressed in stable Hek-DsRed-V96 cells. The images were captured before (10 °C) or after (37 °C) GEPM assembly. Scale bar = 5 µm or 10 µm. b–e) Scatter plots of green and red pixels from converted (8-bit) confocal images. Linear regression lines are shown in red for 10 °C and 37 °C groups. f) For co-transfected cells, Pearson’s coefficients (PC1, PC2) and overlap coefficients (OC) were estimated. PC1 and OC were generated using JACoP software. PC2 was generated from LSM 510 software. A, B, C and D represent the correlation of DsRed-V96/GFP-V96 at 10 °C, DsRed-V96/GFP-V96 at 37 °C, DsRed-V96/GFP-S48I48 at 10 °C and DsRed-V96/GFP-S48I48 at 37 °C, respectively.
In summary, this communication reports the biosynthesis of three different ELP monoblocks and one diblock and their distinct microdomains assembled after ELP-mediated phase separation. It was confirmed that ELP monoblocks with similar Tt assemble relatively large protein coacervates which could coassemble in vitro; however, these coacervates self-sorted from the nanoparticles assembled by ELP diblock copolymers. It is also possible that some mixtures of ELP monoblocks will also sort; however, our data suggests that when the transition temperatures are matched, ELP molecular weight and sequence yield co-assembly. Most importantly, an intracellular assay demonstrated co-assembly of overexpressed DsRed-V96 and transfected GFP-V96 and self-sorting between DsRed-V96 coacervates and GFP-S48I48 nanoparticles in eukaryotic cells. These encouraging in vitro and intracellular findings demonstrate that ELP gene products can be induced to either spatially sort or co-assemble functional proteins (GFP, DsRed) within the cytosol. For the first time, this simple strategy enables advanced control over the organization of micro-structures in the cytosol, which may promote the development of synthetic organelles.
Experimental Section
Biosynthesis and Characterization of ELPs
The recombinant pET25b(+) vectors with ELP gene insertions were used for ELP expression in BLR (DE3) E. coli. Inverse transition cycling was used to purify ELP samples from the cell lysate. The detailed ELP expression and purification procedures were described in the previous publications of our group.[14,18] The optical density of ELPs (OD 350 nm, temperature gradient of 1 °C min−1) was measured using DU800 UV-Vis spectrophotometer (Beckman Coulter, CA). The temperature at the maximum first derivative of the optical density at 350 nm was defined as ELP transition temperature (Tt). Dynamic light scattering (DLS) was used to estimate the hydrodynamic radii of ELP nanoparticles. Pure ELP samples (25 µM, in PBS) were filtered (20 nm membrane filter, 4 °C) and loaded onto a pre-chilled 384 well plate. A Wyatt Dynapro plate reader (Santa Barbara, CA) was used to measure ELP hydrodynamic radii (1 °C temperature interval). Cryogenic-transmission electron microscopy (Cryo-TEM) and negative staining TEM were used to observe nanoparticle morphology in solution and in the dried-down state respectively. The detailed preparation procedure was described in our previous publication.[14]
In Vitro Co-Assembly and Sorting
ELP samples were labeled with rhodamine (Rho) or carboxyfluorescein (CF) using N-Hydroxysuccinimide (NHS) chemistry. The labeled ELPs were diluted to the required concentrations in each designated group to obtain the same Tt. The two ELPs in each group (Rho and CF labeled) were mixed at 1: 1 (v/v) ratio in a 35 mm glass bottom dish (MatTek, MA) and imaged using a Zeiss LSM 510 Meta NLO confocal microscopy (Thornwood, NY) with an Instec HCS60 temperature control stage (Denver, CO). All images were captured under a Plan-Apochromat 63× oil immersion lens with a working distance of 0.19 mm.
Lentivirus (rLV) Production and Generation of Stable Cell Line Hek-DsRed-V96 E xpressing DsRed-V96
DsRed-V96 gene was inserted into pLVX-N1 lentiviral expression vector (Clontech Laboratories, CA). The construct was then mixed with Lenti-X HT proprietary packaging mix and used with Lentiphos HT transfection system (Clontech Laboratories, CA) to transfect 293T cells for production of VSV-G pseudo-typed, replication-incompetent rLV. The supernatants containing the rLV were collected and concentrated by centrifugation (1500 g, 45 min) and LentiX concentrator (Clontech Laboratories, CA) after 48 h post-transfection. Viral titers were determined using p24 antigen ELISA (Invitrogen, CA). To generate a stable cell line, Hek cells were infected with 104 infectious units/ml rLV encoding DsRed-V96. To enrich the tissue culture, the cells were split at 1:10 and 1:100 ratios and continued to grow in the presence of selective agent puromycin. A few resultant single colonies with bright DsRed fluorescence levels were selected using a pipet tip and allowed for cell culture growth (named Hek-DsRed-V96 cells).
Intracellular Co-Assembly and Sorting
ELP genes were inserted to the downstream of NT-GFP-pcDNA3.1 plasmid (Invitrogen, CA) to produce mammalian GFP-ELP fusions. The GFP-ELP constructs were used to transfect Hek-DsRed-V96 cells in plain Dulbecco's modified Eagle’s medium (DMEM) using Turbofect (Fermentas, MA). The cells were incubated with Turbofect-DNA mixture for 6 h, washed with PBS, and cultured in fresh DMEM for 48 h before confocal imaging.[16] Confocal images were captured using a Zeiss LSM 510 Meta NLO confocal microscopy (Thornwood, NY) with an Instec HCS60 temperature control stage (Denver, CO) under a Plan-Apochromat 63× oil immersion lens with a working distance of 0.19 mm.
Statistical Analysis
Confocal images were converted from RGB-color to 8-bit and analyzed by JACoP in ImageJ (NIH). Scatter plots of the red and green pixels, Pearson’s coefficients and overlap coefficients were obtained from JACoP analysis to evaluate colocalization of two ELP microdomains.[19] The slope and r2 value in the linear regression were used to predict the correlation between red and green pixels in the scatter plots. Software LSM 510 (Thornwood, NY) was also used to acquire Pearson’s coefficients from confocal images of each group with a manually determined threshold of noise.
Supplementary Material
Acknowledgements
This work was made possible by the University of Southern California, the National Institute of Health R21EB012281 to J. A. M., and P30 CA014089 to the Norris Comprehensive Cancer Center, the USC Molecular Imaging Center, the USC Nanobiophysics Core Facility, the Translational Research Laboratory at the School of Pharmacy, the American Cancer Society IRG-58–007–48, the Stop Cancer Foundation, the USC Ming Hsieh Institute, and the USC Whittier Foundation. We thank Drs. Liana Asatryan and Daryl Davies at the Lentiviral core of the USC School of Pharmacy for the production of recombinant lentivirus.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Pu Shi, Department of Pharmacology and Pharmaceutical Sciences, University of Southern California, Los Angeles, CA 90089, USA.
Yi-An Lin, Department of Chemical and Biomolecular Engineering, Johns Hopkins University, Baltimore, MD 21218, USA.
Martha Pastuszka, Department of Pharmacology and Pharmaceutical Sciences, University of Southern California, Los Angeles, CA 90089, USA.
Honggang Cui, Department of Chemical and Biomolecular Engineering, Johns Hopkins University, Baltimore, MD 21218, USA.
J. Andrew MacKay, Email: jamackay@usc.edu, Department of Pharmacology and Pharmaceutical Sciences, University of Southern California, Los Angeles, CA 90089, USA.
References
- 1.van Vliet C, Thomas EC, Merino-Trigo A, Teasdale RD, Gleeson PA. Prog. Biophys. Mol. Bio. 2003;83:1. doi: 10.1016/s0079-6107(03)00019-1. [DOI] [PubMed] [Google Scholar]
- 2.Rothman JE. Nature. 1994;372:55. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 3.Mellman I, Nelson WJ. Nat. Rev. Mol. Cell Bio. 2008;9:833. doi: 10.1038/nrm2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearse BM, Robinson MS. Annu. Rev. Cell Bio. 1990;6:151. doi: 10.1146/annurev.cb.06.110190.001055. [DOI] [PubMed] [Google Scholar]
- 5.Palade G. Science. 1975;189:867. doi: 10.1126/science.189.4206.867-b. [DOI] [PubMed] [Google Scholar]
- 6.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Annu. Rev. Cell Dev. Bio. 2004;20:87. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 7.Sun G, Hsueh PY, Janib SM, Hamm-Alvarez S, Andrew MacKay J. J. Control Release. 2011;155:218. doi: 10.1016/j.jconrel.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mackay JA, Chilkoti A. Int. J. Hyperther. 2008;24:483. doi: 10.1080/02656730802149570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah M, Hsueh PY, Sun G, Chang HY, Janib SM, MacKay JA. Protein Sci. 2012;21:743. doi: 10.1002/pro.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urry DW. J. Phys. Chem. B. 1997;101:11007. [Google Scholar]
- 11.Janib SM, Liu S, Park R, Pastuszka MK, Shi P, Moses AS, Orosco MM, Lin YA, Cui H, Conti PS, Li Z, MacKay JA. Integr. Biol. 2013;5:183. doi: 10.1039/c2ib20169k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreher MR, Simnick AJ, Fischer K, Smith RJ, Patel A, Schmidt M, Chilkoti A. J. Am. Chem. Soc. 2008;130:687. doi: 10.1021/ja0764862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright ER, Conticello VP. Adv. Drug Deliv. Rev. 2002;54:1057. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 14.Shi P, Aluri S, Lin YA, Shah M, Edman-Woolcott M, Dhandhukia J, Cui H, Mackay JA. J. Control Release. 2013 doi: 10.1016/j.jconrel.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janib SM, Pastuszka MF, Aluri S, Folchman-Wagner Z, Hsueh PY, Shi P, Lin YA, Cui H, MacKay JA. Polym. Chem. 2014 doi: 10.1039/C3PY00537B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pastuszka MK, Janib SM, Weitzhandler I, Okamoto CT, Hamm-Alvarez S, Mackay JA. Biomacromolecules. 2012;13:3439. doi: 10.1021/bm301090x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge X, Conley AJ, Brandle JE, Truant R, Filipe CD. J. Am. Chem. Soc. 2009;131:9094. doi: 10.1021/ja902890r. [DOI] [PubMed] [Google Scholar]
- 18.Dhandhukia J, Weitzhandler I, Wang W, MacKay JA. Biomacromolecules. 2013;14:976. doi: 10.1021/bm301558q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolte S, Cordelieres FP. J. Microsc. 2006;224:213. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.