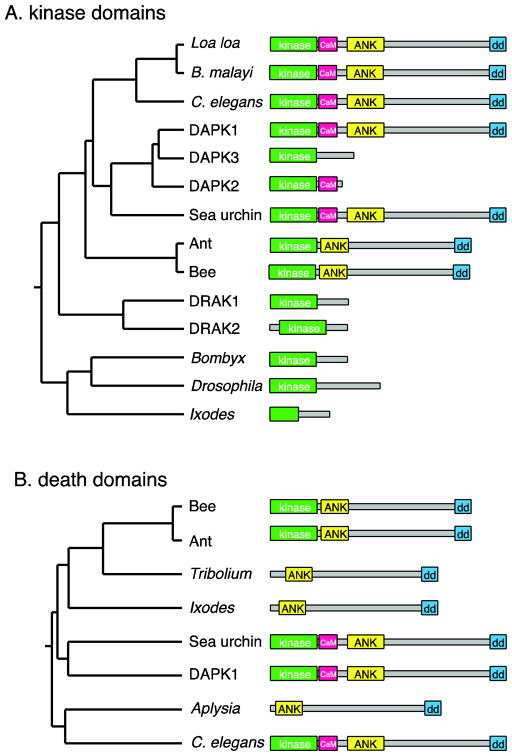
Figure 1. Phylogeny of the DAPK family.
A. Tree based on pairwise alignment (ClustalW) analysis of the amino acid sequences of the kinase domains of DAPK family members, along with cartoons of the protein domain architecture, approximately to scale. Human DAPK1 and DAPK family members are compared with related proteins predicted from genome sequences of various invertebrate species. B: Tree based on alignments of the death domains. To simplify the domain structures, the P-loops and ROCO domains are not shown. NCBI sequence accession numbers: C. elegans (O44997.2); Loa loa (EJD74144.1); Brugia malayi (XP_001896664.1), Human DAPK1 (AAI43760.1); Human DAPK3 (NP_001339.1); Human DAPK2 (Q9UIK4.1); sea urchin Strongylocentrotus purpuratus (XP_003723347.1); carpenter ant Camponotus floridanus (E1ZVV1); honey bee Apis florea (XP_003689717.1); Human DRAK1 (BAA34126.1), Human DRAK2 (BAA34127.1); silkworm Bombyx mori (E9JEH2); Drosophila Drak (E1JJH9); deer tick Ixodes scapularis (EEC12134.1). Kinase-less sequences: flour beetle Tribolium castaneum (XP_972597.2); I. scapularis (EEC05408.1); sea slug Aplysia californica (XP_005108326.1).