Abstract
Background
Gene expression analyses indicate that breast cancer is a heterogeneous disease with at least 5 immunohistologic subtypes. Despite growing evidence that these subtypes are etiologically and prognostically distinct, few studies have investigated whether they have divergent genetic risk factors. To help fill in this gap in our understanding, we examined associations between breast cancer subtypes and previously established susceptibility loci among white and African-American women in the Carolina Breast Cancer Study.
Methods
We used Bayesian polytomous logistic regression to estimate odds ratios (ORs) and 95% posterior intervals (PIs) for the association between each of 78 single nucleotide polymorphisms (SNPs) and 5 breast cancer subtypes. Subtypes were defined using 5 immunohistochemical markers: estrogen receptors (ER), progesterone receptors (PR), human epidermal growth factor receptors 1 and 2 (HER1/2) and cytokeratin (CK) 5/6.
Results
Several SNPs in TNRC9/TOX3 were associated with luminal A (ER/PR+, HER2−) or basal-like breast cancer (ER−, PR−, HER2−, HER1 or CK 5/6+), and one SNP (rs3104746) was associated with both. SNPs in FGFR2 were associated with luminal A, luminal B (ER/PR+, HER2+), or HER2+/ER− disease, but none were associated with basal-like disease. We also observed subtype differences in the effects of SNPs in 2q35, 4p, TLR1, MAP3K1, ESR1, CDKN2A/B, ANKRD16, and ZM1Z1.
Conclusion and Impact
We found evidence that genetic risk factors for breast cancer vary by subtype and further clarified the role of several key susceptibility genes.
Keywords: breast cancer, single nucleotide polymorphisms, breast cancer subtypes, GWAS, Bayesian analysis
INTRODUCTION
Researchers have long recognized that breast cancer is a heterogeneous disease with variable prognoses and clinical characteristics. Further, epidemiologic investigations have discovered evidence of divergent etiologic processes (1, 2), with some key differences in risk factors across disease subgroups (3–6). While these findings have led to advancements in our understanding of the disease, inconsistent subtype definitions and imprecise estimates have hampered progress. Attempts to identify subtype-specific genetic risk factors have been especially discouraging, with little consistency across study populations (7–10).
Most investigators rely on immunohistochemical (IHC) analysis of estrogen receptors (ER), progesterone receptors (PR) and human epidermal growth factor receptors-2 (HER2) to define breast cancer subtypes. These markers are included in most routine clinical evaluations of breast tumors, as they are predictive of response to targeted therapies such as tamoxifen and trastuzumab. Based on concerns that these three markers did not adequately capture disease heterogeneity, researchers turned to gene expression analysis for more in-depth assessments. In one of the first large-scale gene expression analyses of breast tissue, Perou et al. (11) observed that tumors with similar expression patterns also had similar IHC subtypes. The only major exception was triple-negative tumors (i.e. ER−, PR− and HER2−), which clustered into two separate groups with different cytokeratin (CK) 5/6 and human epidermal growth factor receptor-1 (HER1) expression patterns.
This research led to a new classification system with 5 IHC markers serving as adequate, inexpensive surrogates for more complex gene expression profiles (12–14). Because the CK 5/6 protein is usually present in basal epithelial cells but not in more differentiated luminal epithelial cells, the subtypes were designated as follows: Luminal A (ER or PR+, HER2−), Luminal B (ER/PR+, HER2+), HER2+/ER−, and Basal-like (ER, PR−, HER2−, HER1+ or CK 5/6+).
This subtype classification system has led to insights into racial disparities and furthered understanding of etiologic and prognostic differences between disease subgroups. Luminal A is the most common subtype, but subtype prevalence varies by age and race (14–17). Notably, basal-like and other triple-negative tumors are more common in women of African descent (3, 14, 16–20). For women diagnosed before 2000, those with HER2+/ER− and basal-like breast cancers had the poorest prognoses (14, 15, 17, 21). The development and FDA approval of trastuzumab has since improved survival rates for women with HER2+ disease, but women with basal-like or other types of triple-negative disease still experience high short-term mortality (22–24). This phenomenon likely explains some of the racial disparity in mortality between US African-Americans and whites (30.5 versus 21.6 deaths per 100,000 women with breast cancer per year, 2009) (25).
In previous studies of subtype-specific determinants, luminal A breast cancer was associated with most established breast cancer risk factors, including family history of breast cancer, reproductive factors, decreased physical activity, increased alcohol consumption and high breast density (3, 4, 26–41). In case-only risk ratio analyses, women with a family history of the disease, a younger age at diagnosis, or an earlier age at menarche were more likely to have triple-negative than luminal A tumors. Triple-negative tumors were also relatively more common in African-Americans and in women who had more children but did not breastfeed. Risk factors for luminal B and HER2+/ER− subtypes are less established, but evidence suggests that African-American race, family history of breast cancer, lack of breastfeeding and high alcohol consumption are risk factors for HER2+/ER− disease. Luminal B breast cancers are more common in younger women, but otherwise have similar risk profiles to luminal A tumors.
The ground-breaking discovery of the rare but highly penetrant BRCA1 and BRCA2 genes (42, 43) opened a floodgate of linkage analyses, candidate gene studies, and later, genome-wide association studies (GWAS). To date, 74 single nucleotide polymorphisms (SNPs) have met the criteria for genome-wide “discovery” (44) and variants on six candidate genes (ATM, CASP8, CHEK2, CTLA4, NBN, and TP53) have “cumulative evidence of an association” with breast cancer (45). Of the aforementioned variants, only BRCA1 has been consistently linked to a particular subtype, with numerous studies observing associations between BRCA1 mutations and triple-negative disease (12, 46–48) or increased basal marker expression (12, 49).
In an attempt to elucidate subtype-specific genetic risk factors for breast cancer and further our understanding of disease etiology, we estimated associations between breast cancer subtypes and previously identified candidate gene and GWAS hits using women from the Carolina Breast Cancer Study (CBCS). This population is well-suited to answer this research question, as it is one of the few studies to have both a large proportion of African-American participants and information on basal IHC markers.
This evaluation is further enhanced by the use of Bayesian statistical methods. Specifically, based on evidence that most genetic variants have either null or weak associations with breast cancer (44, 45, 50), we improved the precision and overall accuracy of our effect estimates by shrinking them toward an informative, null-centered prior.
MATERIALS AND METHODS
Study population
The CBCS is a population-based, case-control study of invasive and in situ breast cancer. The study was conducted in 24 North Carolina counties between 1993 and 2001. To be eligible, cases had to be between 20 and 74 years of age at the time of their diagnosis, with no prior history of breast cancer. Women with in situ breast cancer were eligible if they were diagnosed with ductal carcinoma in situ with microinvasion to a depth of 2 mm or lobular carcinoma in situ between 1996 and 2001.
Both invasive and in situ cases were identified using the North Carolina Central Cancer Registry’s rapid case ascertainment program (51). A main objective of the CBCS was to collect information on traditionally under-researched populations. Therefore, cases were randomly sampled at disproportionate rates based on race and age. This sampling strategy ensured approximately equal representation of African-American and non-African-American women, as well as younger (age<50) and older women (age 50+).
Throughout the study period, controls aged 20–64 years were selected from North Carolina Department of Motor Vehicles records and were probability matched to cases based on race and age group (52). Controls aged 65–74 were selected form Health Care Financing Administration records in a similar fashion. Women with a history of breast cancer were excluded.
A study nurse conducted detailed in-home interviews of all cases and controls. During the interview, each participant answered questions about her reproductive, medical, and family history, and her exposure to several known or suspected breast cancer risk factors. Each participant was also asked to confirm her age and race and provide a 30 ml blood sample. All participants provided written informed consent and cases were asked to release their medical records and tumor tissue. The Institutional Review Board at the University of North Carolina (UNC) approved this study.
The overall response rate was 77% for cases and 57% for controls. 90% of controls, or 1816 women, provided sufficient blood samples for inclusion in genotype analyses (1105 whites, 681 African-Americans, 30 other race). 88% of cases provided blood samples (2039 women), but only 55% of cases provided both blood and tumor samples (748 whites, 502 African-Americans, 10 other race). This included 247 in situ cases. Individuals who self-identified as a race other than white or African-American were included in overall analyses but excluded from race-specific assessments.
IHC analysis
Tumor tissue and medical records were collected from area hospitals and sent to UNC. ER and PR status was abstracted from the patient’s medical records, when available. If not available, ER and PR IHC assays were performed at the UNC Immunohistochemistry Core Laboratory. Tumors with more than 5% of cells showing nuclei-specific staining were considered receptor positive (53). Agreement between medical records reports and UNC-run assays in 10% random samples of ER+ and ER− tumors was high (concordance = 81%, kappa = 0.62) (14).
All tumor samples with sufficient tissue were assayed for HER2, HER1 and CK 5/6. A case was considered HER2+ if at least 10% of observed cells showed signs of CB11 monoclonal antibody staining (54). Tissue with any sign of cytoplasmic or membranous staining was considered positive for CK 5/6 or HER1, respectively (3, 13). Due to the limited amount of available tissue, in situ staining techniques were slightly modified (see Livasy et al. [1]).
As described above, these subtypes were classified as follows: luminal A (ER+ and/or PR+, HER2−), luminal B (ER+ and/or PR+, HER2+), HER2+/ER− (ER−, PR−, HER2+), and basal-like (ER−, PR−, HER2−, HER1+ and/or CK 5/6+). Additionally, tumors negative for all 5 markers were grouped together as the ‘unclassified’ subtype.
SNP selection
Single nucleotide polymorphisms (SNPs) from ten early breast cancer GWAS (55–62) or GWAS follow-up studies (63, 64) were selected for inclusion in this subtype evaluation study. We included SNPs from these studies that had genome-wide p-values below 10−5 in preliminary or pooled analyses. We also retained SNPs in CASP8, ATM, and TP53, some of the key genes identified in a recent comprehensive meta-analysis (45). Lastly, we included a number of SNPs in the same gene as GWAS selected variants, most of which were originally selected to enhance coverage of these regions. In total, this analysis included 22 GWAS hits, 19 other GWAS-identified variants that fell short of genome-wide significance criteria, 21 SNPs from CASP8, ATM, or TP53, and 21 tag SNPs from select GWAS genes.
Each CBCS participant was genotyped at 144 ancestry informative markers. This genotype information was used to estimate each participant’s proportion of African ancestry. When included in regression models, this ancestry proportion estimate should control confounding due to population stratification (65, 66).
Genotype analysis
The included SNPs were genotyped using either a Taqman panel (Applied Biosystems, Inc.) or a Custom GoldenGate Genotyping assay (Illumina, Inc.). The majority of SNPs were genotyped on the Illumina panel, as described previously (67). The Taqman panel (68) included SNPs that had low Illumina design scores, failed the Illumina assay, or were identified as GWAS hits after the Illumina assays were performed. Eighty-one women with poor genotyping quality on the Illumina panel were assigned missing values for those SNPs. All of the SNPs selected for inclusion in this subtype analysis passed quality control tests, including those for call rate, assay intensity, and genotype clustering.
For each SNP, we examined published studies to determine which allele was associated with an increased risk of breast cancer in previous analyses. This allele was designated as the risk allele. For whites, we selected risk alleles for all ATM, CASP8, and TP53 SNPs based on the Zhang et al. meta-analysis (45). For the remaining SNPs, we ascertained the risk allele in the initial GWAS (54–63) and subsequent replication studies (see supplementary references). In each case, if the 95% confidence interval (CI) limits excluded the null, the odds ratio (OR) for the specified allele was in the same direction as the initial study. Despite some minor discrepancies in the direction of the ORs in African-American only studies, we assigned the same risk allele for both racial groups to allow pooling and facilitate comparisons. For novel SNPs and SNPs with no prior statistically significant findings, we designated the minor variant as the risk variant, using the HapMap CEU population as a reference.
Statistical methods
We calculated case-stratified descriptive statistics for age, proportion of African ancestry, and menopausal status, and then repeated these analyses for white and African-American participants separately. We also examined overall and race-stratified distributions of stage at diagnosis, breast cancer subtype, and ER, PR, and HER2 status. Participants were weighted according to their inverse sampling probability. Similarly, all regression models included an offset term to account for the weighted sampling procedures.
For all SNPs, we calculated overall and race-stratified risk allele frequencies (RAFs). We tested for departures from Hardy-Weinberg equilibrium (HWE) separately in white and African-American controls using Pearson’s chi-squared test. If a SNP had a HWE p-value less than 0.05 in either population, we re-inspected the SNP’s genotype clustering images for indications of poor genotype differentiation or other lab error. As many of the SNPs were located in the same gene or gene regions, we also calculated overall and race-stratified correlation coefficients.
We calculated ORs and 95% posterior intervals (PIs) for the association between each subtype and SNP using Bayesian polytomous logistic regression models. We assumed additive genetic models and adjusted for self-reported race (African-American or non- African-American), proportion of African ancestry, and age at diagnosis or selection. We centered age at 50 years and ancestry at its mean value. We also calculated race-specific ORs and 95% PIs, adjusting for age and ancestry.
Previous studies of the association between known susceptibility variants and breast cancer have produced ORs in the range of 1.1–1.3 (44, 45, 50), but subtype-specific associations are less well-characterized. Bearing this in mind, we assigned each SNP log OR a null-centered prior with a mean of 0, but selected a variance of τ2 ~ 1/Γ(4, 0.5) to reflect the likely effect size. These parameters correspond to prior SNP-subtype ORs with 95% mass between 0.54 and 1.86 when τ2 is equal to the mode of the distribution (0.1). As a full Bayes approach requires priors for all parameters, we also assigned null-centered, lognormal priors for age, ancestry, race and the intercept term. We assigned relatively informative priors to age and ancestry (τ2=0.68), which were both mean-centered variables, but a larger variance to race (τ2=1.0). Because the intercept is difficult to define or interpret in a case-control study with weighted sampling, we assigned a vague prior, with τ2=1000. We assumed that all priors were independent.
Priors were incorporated into regression models using Bayes’ theorem. Briefly, Bayes’ theorem states the posterior probability distribution for the parameter of interest given the observed data, f(β|D), is proportional to the likelihood of the observed data, L(β;D), multiplied by the prior probability distribution f(β) (69–71). The aforementioned likelihood is identical to the likelihood used to obtain standard, frequentist maximum likelihood estimates (MLEs). Put another way, the posterior odds ratio is an inverse-variance weighted combination of the likelihood and prior distribution. Further, the variance of the resulting, normal posterior distribution is the inverse of the sum of the weights.
We also conducted sensitivity analyses, estimating MLE of ORs and 95% CIs and another set of Bayesian ORs and 95% PIs given a more informative, but still null-centered prior [SNP~N(0, τ2), τ2~1/Γ (3, 0.2), mode=0.05]. For each Bayesian model, we took 50,000 samples, discarding the first 1000 draws as a burn in, and thinning by retaining every tenth draw, such that the results are based on 4990 samples. Autocorrelation, trace, and density plots indicated adequate mixing and model convergence. All analyses were conducted using the SAS procedure MCMC (v9.3, Cary, NC). Example code is provided as supplementary material.
RESULTS
As seen in Table 1, white and African-American participants in the CBCS population differed in a few key ways. African-Americans were more likely to have later stage disease, with 63% presenting at stage II or higher, relative to 48% of whites. African-Americans were also less likely to be postmenopausal at the time of their diagnosis and were more likely to have basal-like (22% vs. 11%), unclassified (14% vs. 8%) or HER2+/ER− disease (8% vs. 6%). Luminal A breast cancer was the most common breast cancer subtype overall (60%). Seven SNPs had HWE p-values<0.05 (Table 2), though no SNPs failed HWE tests in both whites and African-Americans. Upon reinspection of genotype clustering images, we found that six of the seven SNPs showed good differentiation with no overlap between genotypes. We excluded the seventh SNP, rs614367 (MYEOV), after discovering evidence of allelic dropout and disparate clustering within the homozygous rare genotype. We also excluded SNPs with minor allele frequencies less than 1% in our sample. This left us with 78 SNPs in the overall analysis, 76 in the white only analysis and 73 in the African-American only analysis. Many of the FGFR2 SNPs were highly correlated with one another, as were the two COX11 SNPs, the two CASP8 SNPs and some TNRC9/TOX3 and ATM SNPs (Supplementary Tables S1a–S1f).
Table 1.
Descriptive statistics for Carolina Breast Cancer Study participants included in subtype analysis
| Cases | Controls | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Overall (%)a N=1260b |
White (%)a N=748 |
African Americans (%)a N=502 |
Overall (%)a N=1816b |
White (%)a N=1105 |
African Americans (%)a N=681 |
|
| Age (years); mean (std) | 51.5 (11.6) | 52.1 (11.8) | 50.8 (11.4) | 52.5 (11.3) | 53.0 (11.2) | 51.9 (11.3) |
| Proportion African Ancestry; mean (std) | 0.35 (0.36) | 0.06 (0.06) | 0.77 (0.13) | 0.33 (0.36) | 0.07 (0.09) | 0.77 (0.14) |
| Postmenopausal; N (%) | 691 (67) | 417 (70) | 269 (56) | 1032 (38) | 640 (37) | 377 (41) |
| Stage of Disease; N (%) | ||||||
| In situ | 247 (10) | 192 (10) | 52 (10) | |||
| Stage I | 369 (39) | 232 (42) | 136 (28) | |||
| Stage II | 492 (42) | 250 (40) | 237 (50) | |||
| Stage III | 100 (7) | 46 (6) | 54 (11) | |||
| Stage IV | 24 (2) | 12 (2) | 11 (2) | |||
| missing | 28 | 16 | 12 | |||
| Subtype; N (%) | ||||||
| Luminal A | 700 (60) | 453 (64) | 242 (49) | |||
| Luminal B | 122 (11) | 82 (11) | 38 (8) | |||
| HER2+/ER− | 98 (6) | 59 (6) | 39 (8) | |||
| Basal-like | 207 (13) | 94 (11) | 112 (22) | |||
| Unclassified | 133 (9) | 60 (8) | 71 (14) | |||
| ER+; N (%) | 745 (66) | 497 (70) | 243 (50) | |||
| PR+; N (%) | 543 (60) | 341 (64) | 197 (45) | |||
| missing | 247 | 192 | 52 | |||
| HER2+; N (%) | 220 (17) | 141 (17) | 77 (16) | |||
Percentages weighted by inverse sampling probability
Includes those who self-identified as a race other than white or African-American
Table 2.
Risk allele frequencies (RAF) by race and case status, African-Americans and non African-Americans in the Carolina Breast Cancer Study
| Gene | Locus | Risk allele | All (1260 cases, 1817 controls)a | Whites (748 cases, 1105 controls) | African-Americans (502 cases, 681 controls) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| RAF casesb | RAF controlsb | RAF casesb | RAF controlsb | HWE p-value | RAF casesb | RAF controlsb | HWE p-value | |||
| 1p12 | rs11249433 | G | 0.37 | 0.36 | 0.44 | 0.41 | 0.54 | 0.14 | 0.10 | 0.01 |
| CASP8 | rs1045485 | G | 0.89 | 0.88 | 0.88 | 0.87 | 0.63 | 0.94 | 0.95 | 0.74 |
| CASP8 | rs17468277 | C | 0.89 | 0.88 | 0.88 | 0.87 | 0.63 | 0.95 | 0.95 | 0.95 |
| 2q35 | rs13387042 | A | 0.59 | 0.52 | 0.55 | 0.47 | 0.83 | 0.73 | 0.73 | 1.00 |
| 2p | rs4666451 | G | 0.64 | 0.65 | 0.60 | 0.63 | 0.30 | 0.78 | 0.77 | 0.12 |
| SLC4A7 | rs4973768 | T | 0.44 | 0.42 | 0.47 | 0.42 | 0.22 | 0.35 | 0.40 | 0.05 |
| 4p | rs12505080 | C | 0.27 | 0.23 | 0.30 | 0.24 | 0.80 | 0.18 | 0.17 | 0.64 |
| TLR1 | rs7696175 | T | 0.38 | 0.38 | 0.46 | 0.45 | 0.91 | 0.09 | 0.06 | 0.54 |
| MRPS30 | rs4415084 | T | 0.46 | 0.45 | 0.42 | 0.42 | 0.18 | 0.64 | 0.58 | 0.70 |
| MRPS30 | rs10941679 | G | 0.27 | 0.28 | 0.29 | 0.30 | 0.76 | 0.19 | 0.19 | 0.17 |
| 5p12 | rs981782 | T | 0.60 | 0.65 | 0.51 | 0.59 | 0.26 | 0.91 | 0.91 | 0.60 |
| 5q | rs30099 | T | 0.11 | 0.10 | 0.10 | 0.10 | 0.40 | 0.15 | 0.12 | 0.75 |
| MAP3K1 | rs889312 | C | 0.33 | 0.35 | 0.33 | 0.34 | 0.85 | 0.33 | 0.36 | 0.08 |
| ESR1 | rs2046210 | A | 0.43 | 0.40 | 0.38 | 0.35 | 0.48 | 0.62 | 0.61 | 0.15 |
| ESR1 | rs851974 | G | 0.36 | 0.39 | 0.41 | 0.43 | 0.28 | 0.18 | 0.17 | 0.46 |
| ESR1 | rs2077647 | A | 0.50 | 0.49 | 0.50 | 0.49 | 0.64 | 0.51 | 0.51 | 0.16 |
| ESR1 | rs2234693 | T | 0.50 | 0.55 | 0.51 | 0.57 | 0.45 | 0.46 | 0.48 | 0.63 |
| ESR1 | rs1801132 | C | 0.79 | 0.78 | 0.76 | 0.76 | 0.43 | 0.90 | 0.88 | 0.36 |
| ESR1 | rs3020314 | C | 0.44 | 0.41 | 0.38 | 0.34 | 0.15 | 0.68 | 0.71 | 0.75 |
| ESR1 | rs3798577 | T | 0.52 | 0.53 | 0.50 | 0.53 | 0.43 | 0.58 | 0.54 | 0.27 |
| ECHDC1 | rs2180341 | G | 0.26 | 0.28 | 0.24 | 0.27 | 0.55 | 0.32 | 0.33 | 0.83 |
| RELN | rs17157903 | T | 0.14 | 0.12 | 0.14 | 0.12 | 0.06 | 0.11 | 0.10 | 0.08 |
| 8q24 | rs13281615 | G | 0.43 | 0.42 | 0.43 | 0.42 | 0.17 | 0.43 | 0.43 | 0.58 |
| 8q24 | rs1562430 | T | 0.59 | 0.56 | 0.60 | 0.57 | 0.78 | 0.53 | 0.53 | 0.61 |
| CDKN2A/B | rs3731257 | T | 0.20 | 0.21 | 0.23 | 0.23 | 0.24 | 0.08 | 0.11 | 0.89 |
| CDKN2A/B | rs3731249 | A | 0.02 | 0.02 | 0.03 | 0.03 | 0.90 | 0.01 | 0.00 | 0.95 |
| CDKN2A/B | rs518394 | G | 0.36 | 0.41 | 0.44 | 0.48 | 0.17 | 0.09 | 0.08 | 0.06 |
| CDKN2A/B | rs564398 | G | 0.35 | 0.40 | 0.42 | 0.47 | 0.29 | 0.08 | 0.08 | 0.02 |
| CDKN2A/B | rs1011970 | T | 0.22 | 0.18 | 0.19 | 0.15 | 0.62 | 0.33 | 0.34 | 0.14 |
| CDKN2A/B | rs10757278 | A | 0.60 | 0.60 | 0.55 | 0.55 | 0.18 | 0.80 | 0.82 | 0.77 |
| CDKN2A/B | rs10811661 | C | 0.14 | 0.18 | 0.16 | 0.20 | 0.02 | 0.07 | 0.07 | 0.24 |
| ANKRD16 | rs2380205 | C | 0.54 | 0.58 | 0.57 | 0.60 | 0.88 | 0.41 | 0.46 | 0.72 |
| ZNF365 | rs10995190 | G | 0.85 | 0.83 | 0.86 | 0.82 | 0.76 | 0.82 | 0.83 | 0.90 |
| ZMIZ1 | rs704010 | T | 0.35 | 0.36 | 0.42 | 0.42 | 0.93 | 0.11 | 0.08 | 0.82 |
| FGFR2 | rs1896395 | A | 0.05 | 0.04 | 0.00 | 0.00 | 0.96 | 0.21 | 0.20 | 0.04 |
| FGFR2 | rs3750817 | C | 0.71 | 0.65 | 0.65 | 0.60 | 0.16 | 0.91 | 0.88 | 0.83 |
| FGFR2 | rs10736303 | G | 0.61 | 0.55 | 0.54 | 0.49 | 0.19 | 0.86 | 0.84 | 0.75 |
| FGFR2 | rs11200014 | A | 0.40 | 0.38 | 0.45 | 0.41 | 0.65 | 0.22 | 0.21 | 0.75 |
| FGFR2 | rs2981579 | T | 0.49 | 0.45 | 0.46 | 0.41 | 0.51 | 0.61 | 0.61 | 0.10 |
| FGFR2 | rs1078806 | G | 0.40 | 0.38 | 0.45 | 0.41 | 0.53 | 0.22 | 0.21 | 0.99 |
| FGFR2 | rs2981578 | C | 0.61 | 0.55 | 0.54 | 0.49 | 0.09 | 0.86 | 0.84 | 0.45 |
| FGFR2 | rs1219648 | G | 0.44 | 0.40 | 0.44 | 0.39 | 0.35 | 0.47 | 0.41 | 0.57 |
| FGFR2 | rs2912774 | A | 0.47 | 0.42 | 0.44 | 0.40 | 0.26 | 0.58 | 0.55 | 0.07 |
| FGFR2 | rs2936870 | T | 0.47 | 0.43 | 0.44 | 0.40 | 0.25 | 0.59 | 0.56 | 0.14 |
| FGFR2 | rs2420946 | T | 0.45 | 0.41 | 0.43 | 0.39 | 0.21 | 0.54 | 0.52 | 0.03 |
| FGFR2 | rs2162540 | G | 0.45 | 0.41 | 0.42 | 0.39 | 0.28 | 0.54 | 0.52 | 0.41 |
| FGFR2 | rs2981582 | T | 0.44 | 0.40 | 0.42 | 0.39 | 0.30 | 0.50 | 0.49 | 0.96 |
| FGFR2 | rs3135718 | G | 0.46 | 0.42 | 0.43 | 0.39 | 0.23 | 0.58 | 0.54 | 0.65 |
| 10q | rs10510126 | C | 0.89 | 0.90 | 0.89 | 0.89 | 0.38 | 0.89 | 0.90 | 0.21 |
| ATM | rs1800054 | G | 0.02 | 0.01 | 0.02 | 0.02 | 0.34 | 0.00 | 0.00 | 0.94 |
| ATM | rs4986761 | C | 0.01 | 0.01 | 0.01 | 0.01 | 0.68 | 0.00 | 0.00 | 0.98 |
| ATM | rs1800056 | C | 0.02 | 0.01 | 0.02 | 0.01 | 0.67 | 0.00 | 0.00 | 0.95 |
| ATM | rs1800057 | G | 0.03 | 0.02 | 0.04 | 0.02 | 0.90 | 0.01 | 0.01 | 0.91 |
| ATM | rs1800058 | T | 0.02 | 0.02 | 0.02 | 0.02 | 0.06 | 0.01 | 0.01 | 0.91 |
| ATM | rs1801516 | A | 0.12 | 0.12 | 0.15 | 0.14 | 0.17 | 0.03 | 0.02 | 0.48 |
| ATM | rs3092992 | C | 0.04 | 0.04 | 0.05 | 0.04 | 0.13 | 0.01 | 0.01 | 0.77 |
| ATM | rs664143 | C | 0.60 | 0.61 | 0.58 | 0.60 | 0.70 | 0.67 | 0.68 | 0.45 |
| ATM | rs170548 | G | 0.27 | 0.33 | 0.32 | 0.37 | 0.88 | 0.09 | 0.12 | 0.07 |
| ATM | rs3092993 | A | 0.12 | 0.12 | 0.15 | 0.14 | 0.19 | 0.03 | 0.02 | 0.48 |
| LSP1 | rs3817198 | C | 0.29 | 0.31 | 0.32 | 0.34 | 0.18 | 0.17 | 0.17 | 0.16 |
| LSP1 | rs909116 | T | 0.57 | 0.56 | 0.53 | 0.52 | 0.20 | 0.72 | 0.72 | 0.96 |
| MYEOV | rs614367 | T | 0.16 | 0.12 | 0.17 | 0.11 | 0.05 | 0.14 | 0.15 | 0.33 |
| H19 | rs2107425 | C | 0.65 | 0.65 | 0.70 | 0.68 | 0.74 | 0.50 | 0.53 | 0.42 |
| TNRC9/TOX3 | rs8049149 | T | 0.00 | 0.00 | 0.00 | 0.00 | 0.98 | 0.01 | 0.02 | 0.32 |
| TNRC9/TOX3 | rs16951186 | T | 0.05 | 0.04 | 0.01 | 0.01 | 0.75 | 0.17 | 0.19 | 0.95 |
| TNRC9/TOX3 | rs8051542 | T | 0.43 | 0.41 | 0.46 | 0.44 | 0.43 | 0.33 | 0.30 | 0.12 |
| TNRC9/TOX3 | rs12443621 | G | 0.50 | 0.43 | 0.51 | 0.41 | 0.39 | 0.48 | 0.51 | 1.00 |
| TNRC9/TOX3 | rs3803662 | T | 0.36 | 0.29 | 0.32 | 0.24 | 0.73 | 0.52 | 0.54 | 0.65 |
| TNRC9/TOX3 | rs4784227 | T | 0.24 | 0.19 | 0.29 | 0.22 | 0.62 | 0.09 | 0.07 | 0.59 |
| TNRC9/TOX3 | rs3104746 | A | 0.08 | 0.05 | 0.03 | 0.02 | 0.48 | 0.26 | 0.18 | 0.87 |
| TNRC9/TOX3 | rs3112562 | G | 0.28 | 0.25 | 0.22 | 0.20 | 0.45 | 0.51 | 0.46 | 0.88 |
| TNRC9/TOX3 | rs9940048 | A | 0.26 | 0.25 | 0.25 | 0.24 | 0.50 | 0.31 | 0.30 | 0.64 |
| TP53 | rs9894946 | G | 0.84 | 0.87 | 0.81 | 0.86 | 0.48 | 0.96 | 0.96 | 0.25 |
| TP53 | rs1614984 | T | 0.41 | 0.39 | 0.41 | 0.39 | 0.22 | 0.39 | 0.40 | 0.03 |
| TP53 | rs4968187 | T | 0.00 | 0.00 | 0.00 | 0.00 | 0.93 | 0.01 | 0.00 | 0.92 |
| TP53 | rs12951053 | C | 0.08 | 0.07 | 0.08 | 0.06 | 0.47 | 0.11 | 0.11 | 0.09 |
| TP53 | rs17880604 | C | 0.01 | 0.01 | 0.01 | 0.01 | 0.21 | 0.00 | 0.00 | 0.95 |
| TP53 | rs1800372 | G | 0.01 | 0.01 | 0.02 | 0.02 | 0.54 | 0.01 | 0.00 | 0.98 |
| TP53 | rs2909430 | G | 0.17 | 0.14 | 0.14 | 0.13 | 0.66 | 0.28 | 0.24 | 0.64 |
| TP53 | rs1042522 | C | 0.67 | 0.71 | 0.75 | 0.77 | 0.64 | 0.40 | 0.43 | 0.77 |
| TP53 | rs8079544 | C | 0.94 | 0.94 | 0.96 | 0.95 | 1.00 | 0.89 | 0.89 | 0.83 |
| COX11 | rs7222197 | G | 0.70 | 0.73 | 0.72 | 0.75 | 0.60 | 0.66 | 0.65 | 0.70 |
| COX11 | rs6504950 | G | 0.70 | 0.73 | 0.72 | 0.75 | 0.59 | 0.66 | 0.65 | 0.66 |
Includes individuals who identified as a race other than white or African-American
Weighted by inverse sampling probability
Several SNPs were associated with luminal A breast cancer, including 13 of 14 evaluated FGFR2 SNPs (ORs≈1.25) and several SNPs in TNRC9/TOX3 (Table 3). The strongest association was seen for rs3104746 on TNRC9/TOX3 (OR=1.58, 95% PI: 1.24, 1.94). Other noteworthy associations included rs13387042 (2q35), rs12505080 (4p), rs7696175 (TLR1), rs889312 (MAP3K1), rs851974 (ESR1), rs1011970 (CDKN2A/B), and rs9894946 (TP53). Many of the previously identified GWAS hits were positively associated with luminal A disease (Figure 1).
Table 3.
Odds ratios and 95% posterior intervals for the association between the selected single nucleotide polymorphisms (SNPs) and each breast cancer subtype, relative to controls [SNP log OR~N(0,τ2), τ2 ~ Γ−1(4, 0.5) with mode=0.10]a
| Gene | SNP | Luminal A N=700 |
Luminal B N=122 |
HER2+/ER− N=98 |
Unclassified N=133 |
Basal-like N=207 |
|---|---|---|---|---|---|---|
| 1p12 | rs11249433 | 1.09 (0.94, 1.25) | 1.04 (0.78, 1.33) | 1.11 (0.79, 1.45) | 1.24 (0.93, 1.58) | 1.09 (0.85, 1.35) |
| CASP8 | rs1045485 | 1.10 (0.88, 1.32) | 1.00 (0.67, 1.35) | 0.97 (0.62, 1.33) | 1.24 (0.80, 1.73) | 1.15 (0.80, 1.56) |
| CASP8 | rs17468277 | 1.11 (0.89, 1.35) | 0.96 (0.64, 1.34) | 0.94 (0.60, 1.32) | 1.29 (0.84, 1.82) | 1.10 (0.79, 1.47) |
| 2q35 | rs13387042 | 1.18 (1.04, 1.35) | 0.99 (0.76, 1.25) | 0.98 (0.69, 1.25) | 1.10 (0.85, 1.40) | 0.92 (0.73, 1.12) |
| 2p | rs4666451 | 0.97 (0.85, 1.11) | 1.03 (0.76, 1.29) | 1.14 (0.84, 1.48) | 1.24 (0.93, 1.56) | 1.16 (0.91, 1.41) |
| SLC4A7 | rs4973768 | 0.98 (0.86, 1.09) | 1.01 (0.78, 1.27) | 0.91 (0.68, 1.17) | 0.93 (0.70, 1.15) | 0.99 (0.78, 1.19) |
| 4p | rs12505080 | 1.18 (1.02, 1.36) | 1.14 (0.84, 1.45) | 1.15 (0.82, 1.54) | 0.88 (0.61, 1.13) | 0.98 (0.75, 1.21) |
| TLR1 | rs7696175 | 1.20 (1.02, 1.38) | 0.99 (0.74, 1.29) | 0.79 (0.54, 1.04) | 1.08 (0.77, 1.38) | 1.27 (0.96, 1.59) |
| MRPS30 | rs4415084 | 1.11 (0.96, 1.25) | 1.06 (0.80, 1.32) | 1.15 (0.85, 1.49) | 1.08 (0.84, 1.35) | 1.17 (0.93, 1.41) |
| MRPS30 | rs10941679 | 1.10 (0.93, 1.27) | 0.94 (0.69, 1.22) | 1.04 (0.73, 1.36) | 0.95 (0.69, 1.23) | 1.21 (0.95, 1.45) |
| 5p12 | rs981782 | 0.90 (0.78, 1.03) | 0.88 (0.64, 1.12) | 1.08 (0.78, 1.41) | 0.99 (0.72, 1.28) | 1.07 (0.82, 1.33) |
| 5q | rs30099 | 1.09 (0.89, 1.31) | 1.03 (0.66, 1.37) | 1.13 (0.75, 1.56) | 0.91 (0.60, 1.20) | 0.99 (0.73, 1.28) |
| MAP3K1 | rs889312 | 1.17 (1.02, 1.33) | 1.04 (0.80, 1.32) | 1.00 (0.74, 1.30) | 1.08 (0.82, 1.35) | 0.89 (0.70, 1.09) |
| ESR1 | rs2046210 | 1.03 (0.90, 1.16) | 1.07 (0.82, 1.36) | 1.29 (0.97, 1.67) | 1.15 (0.88, 1.44) | 1.29 (1.01, 1.55) |
| ESR1 | rs851974 | 0.89 (0.77, 1.02) | 1.13 (0.84, 1.43) | 0.86 (0.62, 1.12) | 0.88 (0.64, 1.10) | 0.96 (0.76, 1.18) |
| ESR1 | rs2077647 | 0.95 (0.83, 1.08) | 1.00 (0.76, 1.25) | 0.98 (0.72, 1.26) | 0.94 (0.71, 1.16) | 0.96 (0.76, 1.17) |
| ESR1 | rs2234693 | 0.91 (0.79, 1.04) | 0.96 (0.75, 1.20) | 1.02 (0.75, 1.27) | 0.86 (0.67, 1.08) | 0.99 (0.79, 1.18) |
| ESR1 | rs1801132 | 1.09 (0.92, 1.28) | 0.91 (0.66, 1.20) | 0.98 (0.67, 1.31) | 1.02 (0.74, 1.32) | 0.85 (0.64, 1.05) |
| ESR1 | rs3020314 | 1.02 (0.89, 1.17) | 1.23 (0.93, 1.54) | 1.08 (0.80, 1.37) | 0.99 (0.77, 1.23) | 1.12 (0.90, 1.34) |
| ESR1 | rs3798577 | 0.97 (0.86, 1.10) | 1.05 (0.79, 1.32) | 0.82 (0.62, 1.02) | 1.10 (0.85, 1.35) | 1.00 (0.82, 1.21) |
| ECHDC1 | rs2180341 | 0.99 (0.84, 1.13) | 1.01 (0.75, 1.31) | 1.04 (0.74, 1.36) | 0.95 (0.70, 1.20) | 1.01 (0.80, 1.23) |
| RELN | rs17157903 | 1.00 (0.83, 1.18) | 1.07 (0.74, 1.41) | 0.77 (0.48, 1.09) | 0.99 (0.70, 1.32) | 1.09 (0.78, 1.39) |
| 8q24 | rs13281615 | 1.04 (0.90, 1.17) | 0.99 (0.75, 1.25) | 1.13 (0.85, 1.43) | 1.08 (0.85, 1.35) | 0.98 (0.79, 1.17) |
| 8q24 | rs1562430 | 1.06 (0.93, 1.19) | 0.96 (0.72, 1.22) | 1.20 (0.90, 1.55) | 1.06 (0.83, 1.32) | 1.07 (0.85, 1.28) |
| CDKN2A/B | rs3731257 | 0.92 (0.78, 1.06) | 0.89 (0.64, 1.17) | 0.96 (0.66, 1.26) | 0.91 (0.66, 1.19) | 0.91 (0.70, 1.16) |
| CDKN2A/B | rs3731249 | 0.99 (0.65, 1.33) | 1.08 (0.55, 1.68) | 1.11 (0.56, 1.76) | 0.95 (0.49, 1.46) | 0.96 (0.51, 1.48) |
| CDKN2A/B | rs518394 | 0.99 (0.86, 1.13) | 1.00 (0.73, 1.26) | 0.77 (0.52, 1.00) | 1.04 (0.76, 1.34) | 1.14 (0.88, 1.38) |
| CDKN2A/B | rs564398 | 1.01 (0.85, 1.15) | 1.01 (0.76, 1.28) | 0.81 (0.56, 1.08) | 1.08 (0.80, 1.40) | 1.09 (0.83, 1.36) |
| CDKN2A/B | rs1011970 | 1.12 (0.96, 1.29) | 0.91 (0.65, 1.15) | 0.90 (0.63, 1.15) | 0.87 (0.63, 1.10) | 1.12 (0.86, 1.38) |
| CDKN2A/B | rs10757278 | 1.05 (0.90, 1.18) | 1.06 (0.80, 1.35) | 1.02 (0.75, 1.31) | 1.22 (0.93, 1.52) | 1.10 (0.86, 1.34) |
| CDKN2A/B | rs10811661 | 0.95 (0.78, 1.13) | 1.01 (0.71, 1.35) | 0.82 (0.54, 1.15) | 0.84 (0.56, 1.12) | 1.10 (0.81, 1.39) |
| ANKRD16 | rs2380205 | 1.03 (0.90, 1.15) | 1.05 (0.79, 1.30) | 1.10 (0.82, 1.40) | 0.80 (0.61, 0.99) | 0.94 (0.77, 1.13) |
| ZNF365 | rs10995190 | 1.03 (0.84, 1.22) | 0.83 (0.59, 1.09) | 0.83 (0.59, 1.11) | 1.24 (0.88, 1.64) | 0.95 (0.74, 1.21) |
| ZMIZ1 | rs704010 | 1.09 (0.94, 1.25) | 1.34 (0.96, 1.70) | 1.36 (0.97, 1.77) | 0.96 (0.70, 1.25) | 1.34 (1.03, 1.66) |
| FGFR2 | rs1896395 | 1.05 (0.80, 1.30) | 0.92 (0.54, 1.33) | 1.31 (0.73, 1.89) | 1.06 (0.70, 1.44) | 1.06 (0.71, 1.40) |
| FGFR2 | rs3750817 | 1.33 (1.13, 1.53) | 1.27 (0.92, 1.64) | 1.22 (0.86, 1.60) | 1.26 (0.92, 1.64) | 1.01 (0.79, 1.25) |
| FGFR2 | rs10736303 | 1.32 (1.14, 1.52) | 1.31 (0.98, 1.65) | 1.26 (0.90, 1.64) | 1.37 (0.99, 1.77) | 0.99 (0.78, 1.22) |
| FGFR2 | rs11200014 | 1.26 (1.10, 1.43) | 1.05 (0.80, 1.31) | 1.19 (0.86, 1.52) | 1.41 (1.08, 1.77) | 0.88 (0.70, 1.07) |
| FGFR2 | rs2981579 | 1.26 (1.10, 1.42) | 1.11 (0.84, 1.41) | 1.34 (1.00, 1.68) | 1.44 (1.11, 1.80) | 0.92 (0.74, 1.10) |
| FGFR2 | rs1078806 | 1.24 (1.08, 1.42) | 1.07 (0.79, 1.36) | 1.19 (0.86, 1.53) | 1.40 (1.06, 1.76) | 0.87 (0.68, 1.07) |
| FGFR2 | rs2981578 | 1.33 (1.15, 1.51) | 1.31 (1.01, 1.67) | 1.34 (0.98, 1.76) | 1.38 (1.03, 1.84) | 1.01 (0.79, 1.25) |
| FGFR2 | rs1219648 | 1.29 (1.13, 1.47) | 1.10 (0.83, 1.37) | 1.24 (0.89, 1.59) | 1.62 (1.24, 2.04) | 0.95 (0.76, 1.15) |
| FGFR2 | rs2912774 | 1.26 (1.10, 1.41) | 1.11 (0.84, 1.39) | 1.42 (1.06, 1.80) | 1.47 (1.14, 1.85) | 0.92 (0.73, 1.09) |
| FGFR2 | rs2936870 | 1.26 (1.09, 1.41) | 1.13 (0.86, 1.43) | 1.38 (1.02, 1.76) | 1.50 (1.16, 1.89) | 0.91 (0.72, 1.09) |
| FGFR2 | rs2420946 | 1.22 (1.06, 1.38) | 1.06 (0.80, 1.32) | 1.40 (1.04, 1.79) | 1.46 (1.12, 1.83) | 0.89 (0.71, 1.07) |
| FGFR2 | rs2162540 | 1.28 (1.11, 1.45) | 1.08 (0.82, 1.36) | 1.42 (1.05, 1.83) | 1.52 (1.17, 1.90) | 0.91 (0.72, 1.10) |
| FGFR2 | rs2981582 | 1.27 (1.09, 1.43) | 1.10 (0.87, 1.40) | 1.39 (1.02, 1.76) | 1.28 (1.00, 1.57) | 0.92 (0.73, 1.10) |
| FGFR2 | rs3135718 | 1.26 (1.10, 1.42) | 1.13 (0.85, 1.40) | 1.35 (1.01, 1.71) | 1.51 (1.17, 1.90) | 0.91 (0.73, 1.09) |
| 10q | rs10510126 | 1.09 (0.88, 1.30) | 1.07 (0.73, 1.46) | 1.08 (0.72, 1.49) | 0.94 (0.63, 1.26) | 1.14 (0.81, 1.48) |
| ATM | rs1800054 | 1.10 (0.66, 1.59) | 1.10 (0.54, 1.76) | 1.12 (0.51, 1.78) | 1.00 (0.49, 1.62) | 1.20 (0.58, 1.91) |
| ATM | rs1800057 | 1.19 (0.81, 1.64) | 0.95 (0.49, 1.48) | 1.33 (0.68, 2.13) | 1.01 (0.50, 1.60) | 1.10 (0.56, 1.67) |
| ATM | rs1800058 | 1.05 (0.67, 1.44) | 0.94 (0.45, 1.46) | 1.03 (0.51, 1.67) | 0.98 (0.47, 1.56) | 0.98 (0.49, 1.56) |
| ATM | rs1801516 | 1.03 (0.82, 1.24) | 1.05 (0.70, 1.43) | 0.97 (0.64, 1.37) | 0.95 (0.63, 1.32) | 0.99 (0.70, 1.32) |
| ATM | rs3092992 | 0.97 (0.68, 1.30) | 0.99 (0.57, 1.49) | 1.19 (0.62, 1.85) | 1.05 (0.61, 1.57) | 1.38 (0.79, 1.97) |
| ATM | rs664143 | 1.09 (0.95, 1.23) | 1.00 (0.77, 1.25) | 1.06 (0.79, 1.35) | 0.94 (0.74, 1.17) | 0.98 (0.79, 1.19) |
| ATM | rs170548 | 0.98 (0.84, 1.13) | 0.90 (0.66, 1.16) | 1.00 (0.72, 1.31) | 0.94 (0.69, 1.22) | 1.00 (0.77, 1.24) |
| ATM | rs3092993 | 1.03 (0.83, 1.25) | 1.06 (0.69, 1.48) | 0.94 (0.60, 1.35) | 0.96 (0.61, 1.29) | 1.03 (0.73, 1.37) |
| LSP1 | rs3817198 | 1.02 (0.88, 1.18) | 0.87 (0.62, 1.10) | 1.19 (0.86, 1.52) | 1.21 (0.92, 1.55) | 1.02 (0.79, 1.26) |
| LSP1 | rs909116 | 1.09 (0.94, 1.23) | 0.92 (0.69, 1.15) | 1.23 (0.90, 1.58) | 1.03 (0.79, 1.30) | 1.09 (0.86, 1.32) |
| H19 | rs2107425 | 1.03 (0.90, 1.17) | 0.93 (0.71, 1.17) | 0.99 (0.74, 1.24) | 0.94 (0.72, 1.18) | 1.00 (0.81, 1.19) |
| TNRC9/TOX3 | rs16951186 | 1.02 (0.78, 1.25) | 1.30 (0.79, 1.88) | 0.84 (0.48, 1.21) | 0.83 (0.52, 1.12) | 0.97 (0.67, 1.31) |
| TNRC9/TOX3 | rs8051542 | 1.10 (0.97, 1.24) | 0.96 (0.74, 1.20) | 1.12 (0.84, 1.43) | 1.31 (1.02, 1.64) | 0.84 (0.66, 1.03) |
| TNRC9/TOX3 | rs12443621 | 1.06 (0.94, 1.20) | 0.93 (0.71, 1.16) | 1.21 (0.92, 1.55) | 0.95 (0.74, 1.18) | 1.00 (0.82, 1.21) |
| TNRC9/TOX3 | rs3803662 | 1.16 (1.01, 1.33) | 1.09 (0.83, 1.35) | 1.01 (0.77, 1.29) | 1.13 (0.88, 1.41) | 0.96 (0.78, 1.16) |
| TNRC9/TOX3 | rs4784227 | 1.32 (1.13, 1.54) | 1.09 (0.76, 1.43) | 1.10 (0.76, 1.46) | 1.29 (0.94, 1.67) | 0.90 (0.66, 1.17) |
| TNRC9/TOX3 | rs3104746 | 1.58 (1.24, 1.94) | 1.05 (0.60, 1.50) | 1.31 (0.80, 1.85) | 1.12 (0.76, 1.58) | 1.49 (1.06, 1.98) |
| TNRC9/TOX3 | rs3112562 | 1.07 (0.93, 1.22) | 0.88 (0.62, 1.14) | 1.46 (1.06, 1.87) | 0.80 (0.60, 1.03) | 1.33 (1.06, 1.62) |
| TNRC9/TOX3 | rs9940048 | 1.13 (0.97, 1.28) | 0.84 (0.61, 1.08) | 1.17 (0.86, 1.50) | 0.98 (0.72, 1.25) | 0.91 (0.72, 1.11) |
| TP53 | rs9894946 | 0.86 (0.72, 1.02) | 0.83 (0.59, 1.12) | 1.01 (0.65, 1.36) | 1.05 (0.71, 1.43) | 1.12 (0.81, 1.49) |
| TP53 | rs1614984 | 1.01 (0.88, 1.13) | 0.94 (0.71, 1.16) | 1.01 (0.78, 1.28) | 1.04 (0.79, 1.30) | 1.13 (0.92, 1.36) |
| TP53 | rs12951053 | 0.95 (0.75, 1.18) | 1.32 (0.86, 1.84) | 1.16 (0.71, 1.62) | 1.25 (0.82, 1.73) | 0.99 (0.70, 1.30) |
| TP53 | rs17880604 | 0.87 (0.49, 1.24) | 0.84 (0.35, 1.38) | 1.21 (0.54, 1.94) | 0.96 (0.46, 1.57) | 1.12 (0.57, 1.76) |
| TP53 | rs1800372 | 0.97 (0.55, 1.38) | 1.24 (0.57, 2.00) | 0.93 (0.39, 1.53) | 1.34 (0.59, 2.22) | 1.04 (0.46, 1.71) |
| TP53 | rs2909430 | 1.12 (0.96, 1.31) | 1.09 (0.78, 1.45) | 1.06 (0.73, 1.38) | 0.88 (0.63, 1.13) | 1.10 (0.85, 1.37) |
| TP53 | rs1042522 | 1.03 (0.89, 1.18) | 0.99 (0.73, 1.26) | 0.82 (0.61, 1.06) | 0.94 (0.70, 1.17) | 0.97 (0.77, 1.18) |
| TP53 | rs8079544 | 0.98 (0.78, 1.23) | 1.38 (0.83, 2.07) | 0.76 (0.45, 1.07) | 0.87 (0.59, 1.20) | 1.05 (0.71, 1.44) |
| COX11 | rs7222197 | 1.06 (0.92, 1.22) | 1.08 (0.80, 1.35) | 1.01 (0.74, 1.29) | 1.09 (0.82, 1.37) | 1.05 (0.85, 1.27) |
| COX11 | rs6504950 | 1.05 (0.91, 1.20) | 1.07 (0.80, 1.37) | 1.00 (0.73, 1.28) | 1.08 (0.81, 1.35) | 1.05 (0.84, 1.28) |
Estimates generated using polytomous logistic regression adjusting for age at diagnosis/selection, proportion African ancestry and self-reported race.
Figure 1.
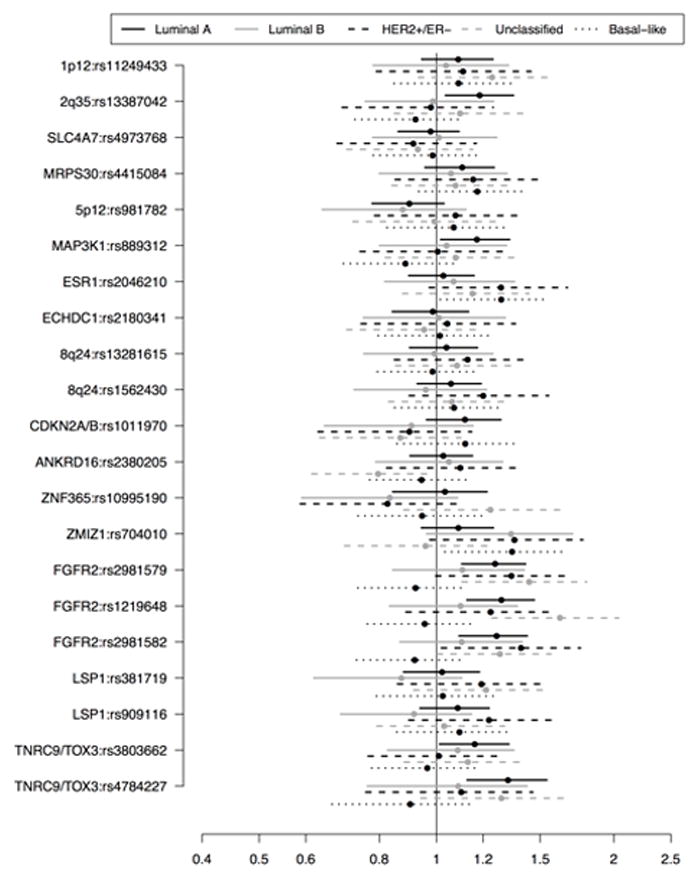
Odds ratios and 95% posterior intervals for previous GWAS–identified SNPs: All CBCS participants
HER2+/ER− disease and unclassified disease were also strongly associated with several FGFR2 and TNRC9/TOX3 SNPs. For both subtypes, the OR estimates for the FGFR2 SNPs were high, with many at or near 1.4. Beyond these key genes, HER2+/ER− disease was positively correlated with the designated risk variant at rs2046210 on ESR1, and rs704010 on ZMIZ1, but negatively correlated with the risk variant at rs7696175 on TLR1, rs3798577 on ESR1, and rs518394 on CDKN2A/B. The ‘C’ allele at rs2380205 (ANKRD16) was inversely associated with the risk of unclassified breast cancer.
We identified relatively few susceptibility variants for luminal B breast cancer. All but one FGFR2 SNP was associated with increased disease risk, but the observed effects were weaker than the other non basal-like subtype ORs, and only one had a posterior interval that excluded the null (rs2981578). The risk allele at rs704010 on ZMIZ1 was also associated with luminal B disease (OR=1.34, 95% PI: 0.96, 1.70).
None of the FGFR2 SNPs were associated with an increased risk of basal-like breast cancer. In fact, most of the FGFR2 ORs for basal-like disease were less than one. Risk variants at two TNRC9/TOX3 SNPs (rs3014746 and rs3112562) were positively associated with basal-like disease, as were risk variants at rs704010 on ZMIZ1 and rs2046210 on ESR1. Additionally, rs7696175 on TLR1 and rs10941679 on MRPS30 each had ORs greater than 1.2 for basal-like breast cancer, relative to controls.
Race-stratified subtype analyses revealed a few additional insights (Supplementary Tables S2 and S3). The most striking was for rs10757278 on CDKN2A/B, where the ‘A’ allele was positively associated with luminal A disease in whites (OR=1.19, 95% PI: 1.02, 1.39) but negatively associated with disease in African-Americans (OR=0.75, 95% PI: 0.58, 0.94). Race-specific results for the previous GWAS hits can be seen in Figure 2.
Figure 2.
Odds ratios and 95% posterior intervals for previous GWAS–identified SNPs for CBCS whites (left) and African–Americans (right)
The two 8q24 SNPs were strongly associated with luminal A breast cancer only among whites (OR=1.16, 95% PI: 0.98, 1.35 and OR=1.17, 95% PI: 1.00, 1.37 for rs13281615 and rs1562430, respectively). The same was true for a TNRC9/TOX3 SNP (rs8051542 OR=1.16, 95% PI: 0.99, 1.35) and a LSP1 SNP (rs909116 OR=1.17, 95% PI: 0.99, 1.37). As for the other subtypes, rs3112562 and rs12443621 (TNRC9/TOX3) were strongly associated with luminal B (OR=0.64, 95% PI: 0.39, 0.89) and HER2+/ER− breast cancer (OR=1.53, 95% PI: 1.05, 2.09), respectively, only among whites. We observed no noteworthy findings in the African-American only analyses.
Results from the MLE analysis and alternate Bayes analysis are presented in Supplementary Tables S4 and S5. Compared with the MLE results, the ORs and PIs presented here are attenuated towards the null and are more precise. As expected, the rarer risk alleles had larger discrepancies between their Bayesian and MLE ORs. For example, the MLE and Bayesian ORs for basal-like breast cancer and rs1800054 (ATM, RAF=1%) were 1.57 (95% CI: 0.66, 3.75) and 1.20 (95% PI: 0.58, 1.91), respectively, compared with MLE and Bayesian ORs of 1.33 (95% CI: 1.02, 1.73) and 1.27 (95% PI: 0.96, 1.59) for rs7696175 (TLR1, RAF=38%). The ORs from the Bayesian analysis with more informative priors were further attenuated. The SNP-subtype association patterns were consistent across all methods.
DISCUSSION
In this study of breast cancer subtypes and previously established susceptibility variants, we observed critical differences in subtype-specific genetic risk factors. The most conspicuous differences involved the FGFR2 gene, where most of the 14 highly correlated SNPs were associated with luminal A, HER2+/ER− and unclassified disease, but not basal-like disease. We also found evidence that SNPs on or near TNRC9/TOX3 are differentially related to breast cancer subtype and that rs10757278 (CDKN2A/B) is differentially related to luminal A disease by race. SNPs in 2q35, 4p, TLR1, MRPS30, MAP3K1, ESR1, ANKRD16, ZM1Z1, and TP53 may also be related to subtype-specific etiology.
As few other studies have employed these enhanced subtype definitions, it is difficult to compare our results with previous reports. Most prior investigations of this topic were limited to comparisons of a single hormone receptor, usually ER+ versus ER− disease (61, 64, 72–82). A few have looked at risk factors for combined ER, PR, HER2 status (7, 9, 10, 83, 84), but to our knowledge, only one other study by Broeks et al. (8) has examined genetic risk factors according to all 5 IHC markers. Broeks et al. (8), Stevens et al. (7), and Han et al. (9) examined some of the SNPs included in this analysis, with some consistencies across populations.
The only FGFR2 SNP examined by Broeks et al. (8) was rs2981582. They also observed positive associations between the T allele and luminal A disease and no association between the SNP and basal-like breast cancer. Their luminal B OR was in the same direction we observed, but of much greater magnitude. Stevens et al. and Han et al. also found near-null associations between rs2981582 and triple negative disease. Contrary to our findings, however, rs2981582 was not associated with HER2+/ER− disease in either study, nor was it associated with unclassified disease in Stevens et al. The effect estimates for rs2981582 and luminal A and B disease reported by Han et al. are similar to those seen in our study. We are the first to report subtype-specific estimates for any other FGFR2 SNPs.
Broeks et al., Stevens et al., and Han et al. also evaluated one TNRC9/TOX3 SNP, rs3803662. Both Broeks et al. and Han et al. observed a positive association with the T allele and luminal A breast cancer, as we did. However, these authors also observed associations between the T allele and luminal B and HER2+/ER− disease, where we found only a weak association with Luminal B and a near-null association with HER2+/ER− disease. Lastly, Broeks et al. observed an association between rs3803662 and basal-like disease, which we did not observe.
These three study groups also assessed other SNPs included in our panel. While it is difficult to draw clear inferences from individual SNP-subtype analyses, these studies, together with ours, suggest that some important differences by subtype do exist. In addition to FGFR2 and TNRC9/TOX3, the effects of rs2046210 (ESR1), rs13387042 (2q35), and rs889312 (MAP3K1) seem to vary according to subtype. Additional studies are needed to further clarify the role of these SNPs and the other potentially important genes identified in our investigation.
While this is one of the first studies to look at genetic risk factors for specific subtypes, breast cancer susceptibility loci are a commonly studied topic. Bayesian methods allowed us to use this plethora of prior information to generate more precise estimates. Assuming we selected reasonable priors, the results presented here will also be more accurate, on average, than those produced using frequentist methods that do not incorporate the wealth of information from prior studies. Further, by selecting null-centered, highly informative priors, bias resulting from these methods is likely to be towards the null (85). In this way, this application of Bayesian methods also reduces the probability of observing false positive associations. We believe the priors specified here are reasonable given existing knowledge of breast cancer susceptibility variants, but we also provide alternate analyses that demonstrate the influence of our assumptions.
Due to differences in blood and tumor sample availability by race, African-Americans were underrepresented in genotyping analyses but overrepresented in IHC analyses. Women with advanced disease were more likely to provide tumor tissue. These trends may result in biased effect estimates for SNPs related to race or disease aggressiveness. Controlling for self-reported race and ancestry should alleviate some of this bias. Though not included in this analysis, we could have used inverse-probability of selection weighting or Bayesian imputation methods to further address this issue.
There is some disagreement in the field as to how best to classify breast cancer subtypes. As discussed, the IHC markers used here are only proxies for more complex gene expression profiles, and thus may not sufficiently capture tumor heterogeneity (86–89). While our approach is likely more informative than one using three or fewer markers, poor subtype specification may attenuate effects and underestimate subtype differences. Misclassification due to inaccurate medical records or IHC evaluations could also bias effects. Other potential sources of misclassification include allelic drop-out and other genotyping errors, though thorough quality control checks likely limited the impact of such errors.
We included in situ cases to increase sample size and improve precision. While this could bias effect estimates of SNPs associated with disease aggressiveness or progression, shared risk profiles (3, 90, 91) and subtype distributions (1, 3, 92) suggest this bias would be small.
The diverse composition of the CBCS population is a major strength of this study. By recruiting a large proportion of African-Americans, study investigators generated a population uniquely suited to answer questions about race and subtype differences in risk factors. To date, this is the largest study to evaluate breast cancer subtypes using a five-marker panel and one of the largest population-based studies of breast cancer in African-Americans.
This analysis of previously established breast cancer susceptibility loci provides strong evidence of etiologic heterogeneity across breast cancer subtypes. Though likely only a small part of the carcinogenic process, the risk variants identified here offer valuable clues about the nature of these diverse pathways. In turn, this vital information may help to advance disease prevention and control efforts.
Supplementary Material
Acknowledgments
We thank the participants of the Carolina Breast Cancer Study, the UNC BioSpecimen Processing Facility for our DNA extractions, blood processing, storage, and sample disbursement (https://genome.unc.edu/bsp), the UNC Mammalian Genotyping Core for CBCS sample genotyping (http://mgc.unc.edu), and Jessica Tse for her technical assistance.
Funding sources: This research was funded in part by the University Cancer Research Fund of North Carolina, the National Cancer Institute Specialized Program of Research Excellence (SPORE) in Breast Cancer (NIH/ NCI P50-CA58223), the Lineberger Comprehensive Cancer Center Core Grant (NIH/NCI P30-CA16086), and an institutional training grant from the National Institute of Health (UNC Lineberger Cancer Control Education Program R25CA057726; K.M. O’Brien).
Abbreviations
- CBCS
Carolina Breast Cancer Study
- CK 5/6
Cytokeratin 5/6
- ER
Estrogen Receptor
- HER1
Human Epidermal Growth Factor Receptor-1
- HER2
Human Epidermal Growth Factor Receptor-2
- HWE
Hardy-Weinberg Equilibrium
- IHC
Immunohistochemical
- MLE
Maximum Likelihood Estimate
- OR
Odds Ratio
- PI
Posterior Interval
- PR
Progesterone Receptor
- RAF
Risk allele frequency
- SNP
Single Nucleotide Polymorphism
- UNC
University of North Carolina
Footnotes
The authors have no financial or nonfinancial conflicts of interest to declare.
References
- 1.Livasy CA, Perou CM, Karaca G, Cowan DW, Maia D, Jackson S, et al. Identification of a basal-like subtype of breast ductal carcinoma in situ. Hum Pathol. 2007;38:197–204. doi: 10.1016/j.humpath.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 2.Melchor L, Benítez J. An integrative hypothesis about the origin and development of sporadic and familial breast cancer subtypes. Carcinogenesis. 2008;29:1475–82. doi: 10.1093/carcin/bgn157. [DOI] [PubMed] [Google Scholar]
- 3.Millikan RC, Newman B, Tse C, Moorman PG, Conway K, Dressler LG, et al. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat. 2008;109:123–39. doi: 10.1007/s10549-007-9632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang XR, Chang-Claude J, Goode EL, Couch FJ, Nevanlinna H, Milne RL, et al. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. J Natl Cancer Inst. 2011;103:250–63. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Althuis MD, Fergenbaum JH, Garcia-Closas M, Brinton LA, Madigan MP, Sherman ME. Etiology of hormone receptor-defined breast cancer: a systematic review of the literature. Cancer Epidemiol Biomarkers Prev. 2004;13:1558–68. [PubMed] [Google Scholar]
- 6.Ma H, Bernstein L, Pike MC, Ursin G. Reproductive factors and breast cancer risk according to joint estrogen and progesterone receptor status: a meta-analysis of epidemiological studies. Breast Cancer Res. 2006;8:R43. doi: 10.1186/bcr1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevens KN, Vachon CM, Lee AM, Slager S, Lesnick T, Olswold C, et al. Common Breast Cancer Susceptibility Loci Are Associated with Triple-Negative Breast Cancer. Cancer Res. 2011;71:6240–6249. doi: 10.1158/0008-5472.CAN-11-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broeks A, Schmidt MK, Sherman ME, Couch FJ, Hopper JL, Dite GS, et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2011;20:3289–303. doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han W, Woo JH, Yu J, Lee M, Moon H, Kang D, et al. Common genetic variants associated with breast cancer in Korean women and differential susceptibility according to intrinsic subtype. Cancer Epidemiol Biomarkers Prev. 2011;20:793–8. doi: 10.1158/1055-9965.EPI-10-1282. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nature Genetics. 2013;45:392–398. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 12.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10:5367–74. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 14.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 15.Cheang MCU, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14:1368–76. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 16.Huo D, Ikpatt F, Khramtsov A, Dangou J, Nanda R, Dignam J, et al. Population differences in breast cancer: survey in indigenous African women reveals over-representation of triple-negative breast cancer. J Clin Oncol. 2009;27:4515–21. doi: 10.1200/JCO.2008.19.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurebayashi J, Moriya T, Ishida T, Hirakawa H, Kurosumi M, Akiyama F, et al. The prevalence of intrinsic subtypes and prognosis in breast cancer patients of different races. Breast. 2007;16 (Suppl 2):S72–7. doi: 10.1016/j.breast.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 18.Lund MJ, Trivers KF, Porter PL, Coates RJ, Leyland-Jones B, Brawley OW, et al. Race and triple negative threats to breast cancer survival: a population-based study in Atlanta, GA. Breast Cancer Res Treat. 2009;113:357–70. doi: 10.1007/s10549-008-9926-3. [DOI] [PubMed] [Google Scholar]
- 19.Parise CA, Bauer KR, Caggiano V. Variation in breast cancer subtypes with age and race/ethnicity. Crit Rev Oncol Hematol. 2010;76:44–52. doi: 10.1016/j.critrevonc.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Stark A, Kleer CG, Martin I, Awuah B, Nsiah-Asare A, Takyi V, et al. African ancestry and higher prevalence of triple-negative breast cancer: findings from an international study. Cancer. 2010;116:4926–32. doi: 10.1002/cncr.25276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto Y, Ibusuki M, Nakano M, Kawasoe T, Hiki R, Iwase H. Clinical significance of basal-like subtype in triple-negative breast cancer. Breast Cancer. 2009;16:260–7. doi: 10.1007/s12282-009-0150-8. [DOI] [PubMed] [Google Scholar]
- 22.Perez EA, Romond EH, Suman VJ, Jeong J, Davidson NE, Geyer CE, Jr, et al. Four-year follow-up of trastuzumab plus adjuvant chemotherapy for operable human epidermal growth factor receptor 2-positive breast cancer: joint analysis of data from NCCTG N9831 and NSABP B-31. J Clin Oncol. 2011;29:3366–73. doi: 10.1200/JCO.2011.35.0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–8. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 24.O’Brien KM, Cole SR, Tse C, Perou CM, Carey LA, Foulkes WD, et al. Intrinsic breast tumor subtypes, race, and long-term survival in the Carolina Breast Cancer Study. Clin Cancer Res. 2010;16:6100–10. doi: 10.1158/1078-0432.CCR-10-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.US Cancer Morality Statistics. [Accessed January 9, 2013]; http://seer.cancer.gov/canques/mortality.html.
- 26.Tamimi RM, Colditz GA, Hazra A, Baer HJ, Hankinson SE, Rosner B, et al. Traditional breast cancer risk factors in relation to molecular subtypes of breast cancer. Breast Cancer Res Treat. 2012;131:159–167. doi: 10.1007/s10549-011-1702-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gierach G, Burke A, Anderson WF. Epidemiology of triple negative breast cancers. Breast Dis. 2010;32:5–24. doi: 10.3233/BD-2010-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang XR, Sherman ME, Rimm DL, Lissowska J, Brinton LA, Peplonska B, et al. Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol Biomarkers Prev. 2007;16:439–43. doi: 10.1158/1055-9965.EPI-06-0806. [DOI] [PubMed] [Google Scholar]
- 29.Gaudet MM, Press MF, Haile RW, Lynch CF, Glaser SL, Schildkraut J, et al. Risk factors by molecular subtypes of breast cancer across a population-based study of women 56 years or younger. Breast Cancer Res Treat. 2011;130:587–597. doi: 10.1007/s10549-011-1616-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phipps AI, Buist DSM, Malone KE, Barlow WE, Porter PL, Kerlikowske K, et al. Family history of breast cancer in first-degree relatives and triple-negative breast cancer risk. Breast Cancer Res Treat. 2011;126:671–8. doi: 10.1007/s10549-010-1148-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phipps AI, Chlebowski RT, Prentice R, McTiernan A, Stefanick ML, Wactawski-Wende J, et al. Body size, physical activity, and risk of triple-negative and estrogen receptor-positive breast cancer. Cancer Epidemiol Biomarkers Prev. 2011;20:454–63. doi: 10.1158/1055-9965.EPI-10-0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phipps AI, Buist DSM, Malone KE, Barlow WE, Porter PL, Kerlikowske K, et al. Reproductive history and risk of three breast cancer subtypes defined by three biomarkers. Cancer Causes Control. 2011;22:399–405. doi: 10.1007/s10552-010-9709-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phipps AI, Chlebowski RT, Prentice R, McTiernan A, Wactawski-Wende J, Kuller LH, et al. Reproductive history and oral contraceptive use in relation to risk of triple-negative breast cancer. J Natl Cancer Inst. 2011;103:470–7. doi: 10.1093/jnci/djr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shinde SS, Forman MR, Kuerer HM, Yan K, Peintinger F, Hunt KK, et al. Higher parity and shorter breastfeeding duration: association with triple-negative phenotype of breast cancer. Cancer. 2010;116:4933–43. doi: 10.1002/cncr.25443. [DOI] [PubMed] [Google Scholar]
- 35.Xing P, Li J, Jin F. A case-control study of reproductive factors associated with subtypes of breast cancer in Northeast China. Med Oncol. 2010;27:926–31. doi: 10.1007/s12032-009-9308-7. [DOI] [PubMed] [Google Scholar]
- 36.Ma H, Wang Y, Sullivan-Halley J, Weiss L, Marchbanks PA, Spirtas R, et al. Use of four biomarkers to evaluate the risk of breast cancer subtypes in the women’s contraceptive and reproductive experiences study. Cancer Res. 2010;70:575–87. doi: 10.1158/0008-5472.CAN-09-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trivers KF, Lund MJ, Porter PL, Liff JM, Flagg EW, Coates RJ, et al. The epidemiology of triple-negative breast cancer, including race. Cancer Causes Control. 2009;20:1071–82. doi: 10.1007/s10552-009-9331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dolle JM, Daling JR, White E, Brinton LA, Doody DR, Porter PL, et al. Risk factors for triple-negative breast cancer in women under the age of 45 years. Cancer Epidemiol Biomarkers Prev. 2009;18:1157–66. doi: 10.1158/1055-9965.EPI-08-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwan ML, Kushi LH, Weltzien E, Maring B, Kutner SE, Fulton RS, et al. Epidemiology of breast cancer subtypes in two prospective cohort studies of breast cancer survivors. Breast Cancer Res. 2009;11:R31. doi: 10.1186/bcr2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stead LA, Lash TL, Sobieraj JE, Chi DD, Westrup JL, Charlot M, et al. Triple-negative breast cancers are increased in black women regardless of age or body mass index. Breast Cancer Res. 2009;11:R18. doi: 10.1186/bcr2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phipps AI, Malone KE, Porter PL, Daling JR, Li CI. Reproductive and hormonal risk factors for postmenopausal luminal, HER-2-overexpressing, and triple-negative breast cancer. Cancer. 2008;113:1521–6. doi: 10.1002/cncr.23786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684–9. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 43.Wooster R, Neuhausen SL, Mangion J, Quirk Y, Ford D, Collins N, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12–13. Science. 1994;265:2088–90. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 44.Hindorff LA, MacArthur J, Morales J, et al. European Bioinformatics Institute. [Accessed January 13, 2013.];A Catalog of Published Genome-Wide Association Studies. Available at: www.genome.gov/gwastudies.
- 45.Zhang B, Beeghly-Fadiel A, Long J, Zheng W. Genetic variants associated with breast-cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Lancet Oncol. 2011;12:477–88. doi: 10.1016/S1470-2045(11)70076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Evans DG, Howell A, Ward D, Lalloo F, Jones JL, Eccles DM. Prevalence of BRCA1 and BRCA2 mutations in triple negative breast cancer. J Med Genet. 2011;48:520–2. doi: 10.1136/jmedgenet-2011-100006. [DOI] [PubMed] [Google Scholar]
- 47.Haffty BG, Yang Q, Reiss M, Kearney T, Higgins SA, Weidhaas J, et al. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol. 2006;24:5652–7. doi: 10.1200/JCO.2006.06.5664. [DOI] [PubMed] [Google Scholar]
- 48.Lee E, McKean-Cowdin R, Ma H, Spicer DV, Van Den Berg D, Bernstein L, et al. Characteristics of Triple-Negative Breast Cancer in Patients With a BRCA1 Mutation: Results From a Population-Based Study of Young Women. J Clin Oncol. 2011;29:4373–4380. doi: 10.1200/JCO.2010.33.6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foulkes WD, Stefansson IM, Chappuis PO, Bégin LR, Goffin JR, Wong N, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95:1482–5. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 50.Hunter DJ. Lessons from genome-wide association studies for epidemiology. Epidemiology. 2012;23:363–7. doi: 10.1097/EDE.0b013e31824da7cc. [DOI] [PubMed] [Google Scholar]
- 51.Newman B, Moorman PG, Millikan R, Qaqish BF, Geradts J, Aldrich TE, et al. The Carolina Breast Cancer Study: integrating population-based epidemiology and molecular biology. Breast Cancer Res Treat. 1995;35:51–60. doi: 10.1007/BF00694745. [DOI] [PubMed] [Google Scholar]
- 52.Weinberg CR, Sandler DP. Randomized recruitment in case-control studies. Am J Epidemiol. 1991;134:421–432. doi: 10.1093/oxfordjournals.aje.a116104. [DOI] [PubMed] [Google Scholar]
- 53.Huang WY, Newman B, Millikan RC, Schell MJ, Hulka BS, Moorman PG. Hormone-related factors and risk of breast cancer in relation to estrogen receptor and progesterone receptor status. Am J Epidemiol. 2000;151:703–14. doi: 10.1093/oxfordjournals.aje.a010265. [DOI] [PubMed] [Google Scholar]
- 54.Millikan R, Eaton A, Worley K, Biscocho L, Hodgson E, Huang W, et al. HER2 codon 655 polymorphism and risk of breast cancer in African Americans and whites. Breast Cancer Res Treat. 2003;79:355–64. doi: 10.1023/a:1024068525763. [DOI] [PubMed] [Google Scholar]
- 55.Long J, Cai Q, Shu X, Qu S, Li C, Zheng Y, et al. Identification of a functional genetic variant at 16q12. 1 for breast cancer risk: results from the Asia Breast Cancer Consortium. PLoS Genet. 2010;6:e1001002. doi: 10.1371/journal.pgen.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–7. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24. 1 (RAD51L1) Nat Genet. 2009;41:579–84. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng W, Long J, Gao Y, Li C, Zheng Y, Xiang Y, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25. 1. Nat Genet. 2009;41:324–8. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gold B, Kirchhoff T, Stefanov S, Lautenberger J, Viale A, Garber J, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22. 33. Proc Natl Acad Sci U S A. 2008;105:4340–5. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–9. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 62.Easton DF, Pooley KA, Dunning AM, Pharoah PDP, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23. 2. Nat Genet. 2009;41:585–90. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, Jonsson GF, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–6. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 65.Thomas DC, Witte JS. Point: population stratification: a problem for case-control studies of candidate-gene associations? Cancer Epidemiol Biomarkers Prev. 2002;11:505–12. [PubMed] [Google Scholar]
- 66.Barnholtz-Sloan JS, McEvoy B, Shriver MD, Rebbeck TR. Ancestry estimation and correction for population stratification in molecular epidemiologic association studies. Cancer Epidemiol Biomarkers Prev. 2008;17:471–7. doi: 10.1158/1055-9965.EPI-07-0491. [DOI] [PubMed] [Google Scholar]
- 67.Nyante SJ, Gammon MD, Kaufman JS, Bensen JT, Lin DY, Barnholtz-Sloan JS, et al. Common genetic variation in adiponectin, leptin, and leptin receptor and association with breast cancer subtypes. Breast Cancer Res Treat. 2011;129:593–606. doi: 10.1007/s10549-011-1517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bortsov AV, Millikan RC, Belfer I, Boortz-Marx RL, Arora H, McLean SA. μ-Opioid receptor gene A118G polymorphism predicts survival in patients with breast cancer. Anesthesiology. 2012;116:896–902. doi: 10.1097/ALN.0b013e31824b96a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Greenland S. Bayesian perspectives for epidemiological research: I. Foundations and basic methods. Int J Epidemiol. 2006;35:765–75. doi: 10.1093/ije/dyi312. [DOI] [PubMed] [Google Scholar]
- 70.Gill J. Bayesian methods: A social and behavioral sciences approach. 2. Boca Raton, FL: Chapman and Hall/CRC Press; 2002. [Google Scholar]
- 71.Cole SR, Chu H, Greenland S, Hamra G, Richardson DB. Bayesian posterior distributions without Markov chains. Am J Epidemiol. 2012;175:368–75. doi: 10.1093/aje/kwr433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Udler MS, Meyer KB, Pooley KA, Karlins E, Struewing JP, Zhang J, et al. FGFR2 variants and breast cancer risk: fine-scale mapping using African American studies and analysis of chromatin conformation. Hum Mol Genet. 2009;18:1692–703. doi: 10.1093/hmg/ddp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim H, Lee J, Sung H, Choi J, Park SK, Lee K, et al. A genome-wide association study identifies a breast cancer risk variant in ERBB4 at 2q34: results from the Seoul Breast Cancer Study. Breast Cancer Res. 2012;14:R56. doi: 10.1186/bcr3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Slattery ML, Baumgartner KB, Giuliano AR, Byers T, Herrick JS, Wolff RK. Replication of five GWAS-identified loci and breast cancer risk among Hispanic and non-Hispanic white women living in the Southwestern United States. Breast Cancer Res Treat. 2011;129:531–9. doi: 10.1007/s10549-011-1498-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Long J, Shu X, Cai Q, Gao Y, Zheng Y, Li G, et al. Evaluation of breast cancer susceptibility loci in Chinese women. Cancer Epidemiol Biomarkers Prev. 2010;19:2357–65. doi: 10.1158/1055-9965.EPI-10-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Campa D, Kaaks R, Le Marchand L, Haiman CA, Travis RC, Berg CD, et al. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. J Natl Cancer Inst. 2011;103:1252–63. doi: 10.1093/jnci/djr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reeves GK, Travis RC, Green J, Bull D, Tipper S, Baker K, et al. Incidence of breast cancer and its subtypes in relation to individual and multiple low-penetrance genetic susceptibility loci. JAMA. 2010;304:426–34. doi: 10.1001/jama.2010.1042. [DOI] [PubMed] [Google Scholar]
- 78.Garcia-Closas M, Hall P, Nevanlinna H, Pooley K, Morrison J, Richesson DA, et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rebbeck TR, DeMichele A, Tran TV, Panossian S, Bunin GR, Troxel AB, et al. Hormone-dependent effects of FGFR2 and MAP3K1 in breast cancer susceptibility in a population-based sample of post-menopausal African-American and European-American women. Carcinogenesis. 2009;30:269–74. doi: 10.1093/carcin/bgn247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Palmer JR, Ruiz-Narvaez EA, Rotimi CN, Cupples LA, Cozier YC, Adams-Campbell LL, et al. Genetic susceptibility Loci for subtypes of breast cancer in an african american population. Cancer Epidemiol Biomarkers Prev. 2013;22:127–34. doi: 10.1158/1055-9965.EPI-12-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lambrechts D, Truong T, Justenhoven C, Humphreys MK, Wang J, Hopper JL, et al. 11q13 is a susceptibility locus for hormone receptor positive breast cancer. Hum Mutat. 2012;33:1123–32. doi: 10.1002/humu.22089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, et al. Large scale genotyping identifies 41 new loci associated with breast cancer risk. Nature Genetics. 2013;45:353–361. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Haiman CA, Chen GK, Vachon CM, Canzian F, Dunning A, Millikan RC, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat Genet. 2011;43:1210–1214. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stevens KN, Fredericksen Z, Vachon CM, Wang X, Margolin S, Lindblom A, et al. 19p13. 1 is a triple-negative-specific breast cancer susceptibility locus. Cancer Res. 2012;72:1795–803. doi: 10.1158/0008-5472.CAN-11-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hamra GB, Maclehose RF, Cole SR. Sensitivity Analyses for Sparse-Data Problems-Using Weakly Informative Bayesian Priors. Epidemiology. 2013;24:233–239. doi: 10.1097/EDE.0b013e318280db1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ma CX, Luo J, Ellis MJ. Molecular Profiling of Triple Negative Breast Cancer. Breast Dis. 2011;32:73–84. doi: 10.3233/BD-2010-0309. [DOI] [PubMed] [Google Scholar]
- 87.Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist. 2011;16 (Suppl 1):61–70. doi: 10.1634/theoncologist.2011-S1-61. [DOI] [PubMed] [Google Scholar]
- 88.Rody A, Karn T, Liedtke C, Pusztai L, Ruckhaeberle E, Hanker L, et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011;13:R97. doi: 10.1186/bcr3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kerlikowske K. Epidemiology of ductal carcinoma in situ. J Natl Cancer Inst Monogr. 2010:139–41. doi: 10.1093/jncimonographs/lgq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Phillips LS, Millikan RC, Schroeder JC, Barnholtz-Sloan JS, Levine BJ. Reproductive and hormonal risk factors for ductal carcinoma in situ of the breast. Cancer Epidemiol Biomarkers Prev. 2009;18:1507–14. doi: 10.1158/1055-9965.EPI-08-0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou W, Jirström K, Johansson C, Amini R, Blomqvist C, Agbaje O, et al. Long-term survival of women with basal-like ductal carcinoma in situ of the breast: a population-based cohort study. BMC Cancer. 2010;10:653. doi: 10.1186/1471-2407-10-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



