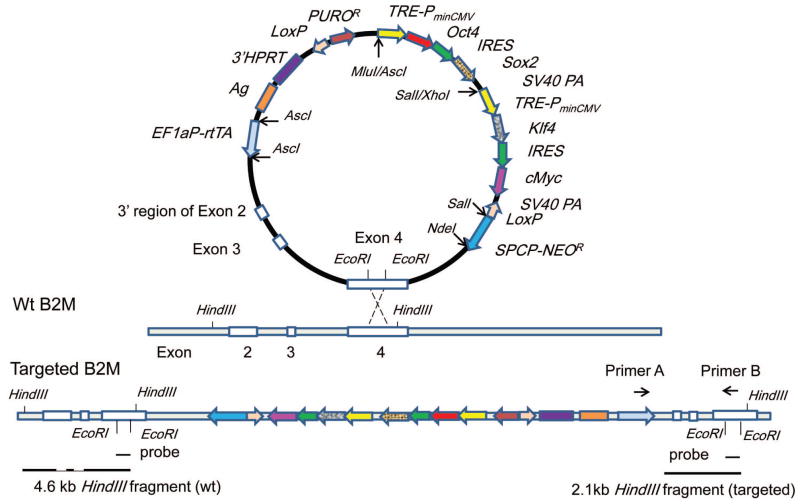
Figure 1. Schematic diagram of 3′hprt.OSMK-LoxP.rtTA.SPCPNEOR.B2M Targeting Vector.
The schematic diagram of the vector is to depict relevant positional information, hence it is not scaled proportionally based on sequence lengths. This vector harbors the reverse tetracycline controlled transactivator (rtTA) under the control of EF-1a promoter, and the four reprogramming factor (Oct4, Sox2, cMyc and Klf4) transgenes under the control of Tet-responsive promoter (TREPminCMV fragment). The two components of Tet-On inducible gene expression system, the regulator (rtTA) and the Tet-responsive promoter in the vector, allow induced expression of Oct4, Sox2, cMyc and Klf4 for hiPSC reprogramming more efficiently. In addition, the LoxP sites in the vector allow removal of puromycin and the four reprogramming factor transgenes by Cre-mediated recombination to generate exogenous reprogramming-factor-free hiPSCs. In order to selectively differentiate hiPSCs into ATIICs, the SPCP-NEOR trangene was added into the targeting vector. One 4.2 kb DNA fragment homologous to the 3′ region of B2M, including partial Exon2, Exon3, Exon4 and 3′ untranslated region, is included in the vector to specifically knock-in all of the transgenes into the 3′ region of B2M gene by homologous recombination. The homologous insertion strategy is expected to result in high targeting frequencies (10–30% on average) and a duplicate of the homologous region contained in the targeting vector [17]. As the insertion targeting vector harbors the 3′ region of B2M gene, the B2M gene should remain intact without causing gene dysfunction after site-specific targeting. To generate hiPSCs, EcoRI was used to linearize the vector and delete a 216 bp gap within Exon 4 of B2M fragment before transfection for gap repair targeting at the B2M gene locus. A 216 bp gap DNA fragment was used as probe, which can hybridize to 4.6 kb wt and 2.1 kb targeted B2M HindIII fragment. In addition, PCR can be performed by using primer A located within rtTA transgene and primer B in the gap region of B2M to identify B2M-targeted transgene.