Figure 1. Sequence determinants of CAL PDZ binding and CAL/NHERF specificity.
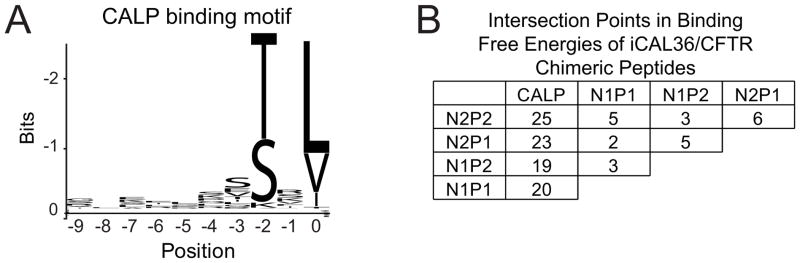
(A) WebLogo analysis (Crooks et al., 2004) of the top 100 sequences that bound CALP in a 6223HumLib peptide array experiment reveals clear preferences at the P0 and P−2 positions, similar to the short, degenerate binding motifs seen with the NHERF PDZ domains (Figure S1A). The C-terminal residue (P0) position is labeled 0; adjacent residues are −1, −2, etc.
(B) For each PDZ domain, free energies of binding (kcal/mol) were calculated from the binding affinities determined by FP for 10 different iCAL36/CFTR chimeric peptide sequences (Table 1). The number of intersection points in pairwise free-energy plots (Figure S2) were tabulated to reveal rank-order exchanges as a function of sequence.
