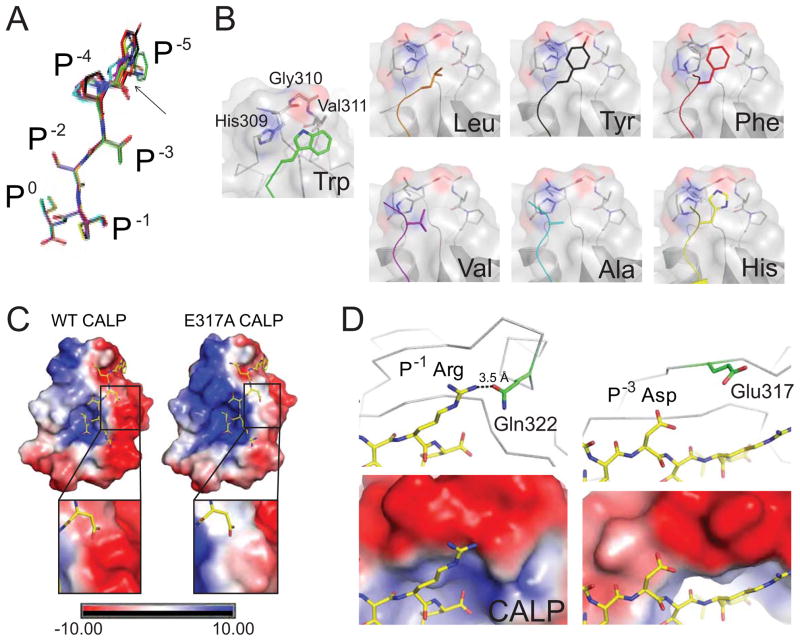
Figure 5. Stereochemical analysis of modulator preferences.
(A) The clustering of CALP-bound P−5-substituted iCAL36 peptides (stick figures) is shown following main-chain superposition of the CALP B-protomers. Peptides are colored by sequence: iCAL36 (W; green carbons), F- (red), Y- (black), H- (yellow), A- (cyan), L- (orange), and V- (purple). RMSD offsets at the P0-P−3 residues were within coordinate error, while modest variations were observed at the P−4 and P−5 positions (black arrow). The largest difference is 3.2 Å, between the Cα of the L- and Cα of the A-substituted peptides.
(B) Each substituted P−5 side chain (colored stick figure) is shown, as docked against its CALP binding interface (surface, cartoon and stick figure). The residues that contact the P−5 residue (His309, Gly310, Val311) are labeled in the CALP:F-iCAL36 structure. Each panel is labeled with the P−5 residue identity. See also Figure S6.
(C) The electrostatic potential surface of WT CALP (left) reveals a highly polarized distribution on either side of the peptide binding cleft. The electrostatic potential surface of CALP-E317A (right) shows a reduced, although still negative, interaction surface for the P−3 Asp residue of iCAL36QDTRL (insets). See also Figure S7.
(D) Electrostatic interactions are shown for P−1 Arg (left) and P−3 Asp (right) residues of the iCAL36QDTRL peptide (stick figure, yellow carbons) bound to CALP (gray Cα trace). At the top, nearby charged CALP residues are shown (stick figures, green carbons). The P−1 Arg forms a hydrogen bond (dashed line) with Gln314. The closets charged residue for the P−3 Asp is Glu317 (d > 5 Å). In the lower panels, the corresponding electrostatic potential surfaces are shown (rendered at 10 kBT/e) underscoring the negative electrostatic environment of both the P−1 and the P−3 side chains.