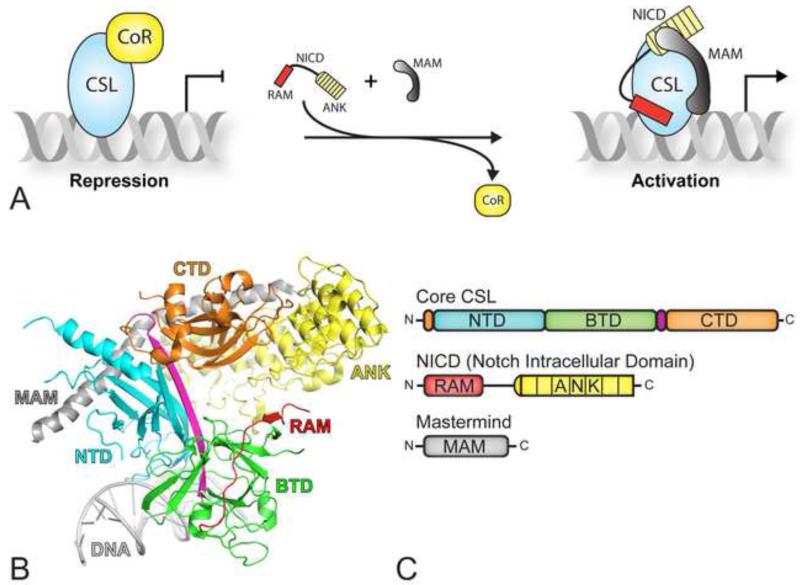
Figure 1. CSL mediated transcriptional regulation.
(A) In the absence of a Notch signal, CSL keeps target genes in a repressed state through interactions with transcriptional corepressors (left). In the presence of a Notch signal, NICD (Notch intracellular domain) translocates to the nucleus, and interacts with CSL and the coactivator Mastermind (MAM), displacing corepressors from CSL and resulting in the activation of Notch target genes. (B) The X-ray structure of CSL in complex with NICD and MAM (PDB ID: 2FO1), referred to as the ternary complex (Wilson and Kovall, 2006). CSL is composed of the N-terminal domain (NTD-cyan), the β-trefoil domain (BTD-green), and the C-terminal domain (CTD-orange). The RAM (RBPJ-Associated Molecule, red) and ANK (ankyrin repeats, yellow) domains of NICD bind the BTD and CTD of CSL, respectively; MAM (grey) forms a long α-helix, which binds along a groove formed by ANK, CTD, and NTD. (C) Domain schematic of core CSL (top), NICD (middle) and MAM (bottom); domains are colored as in panel B.