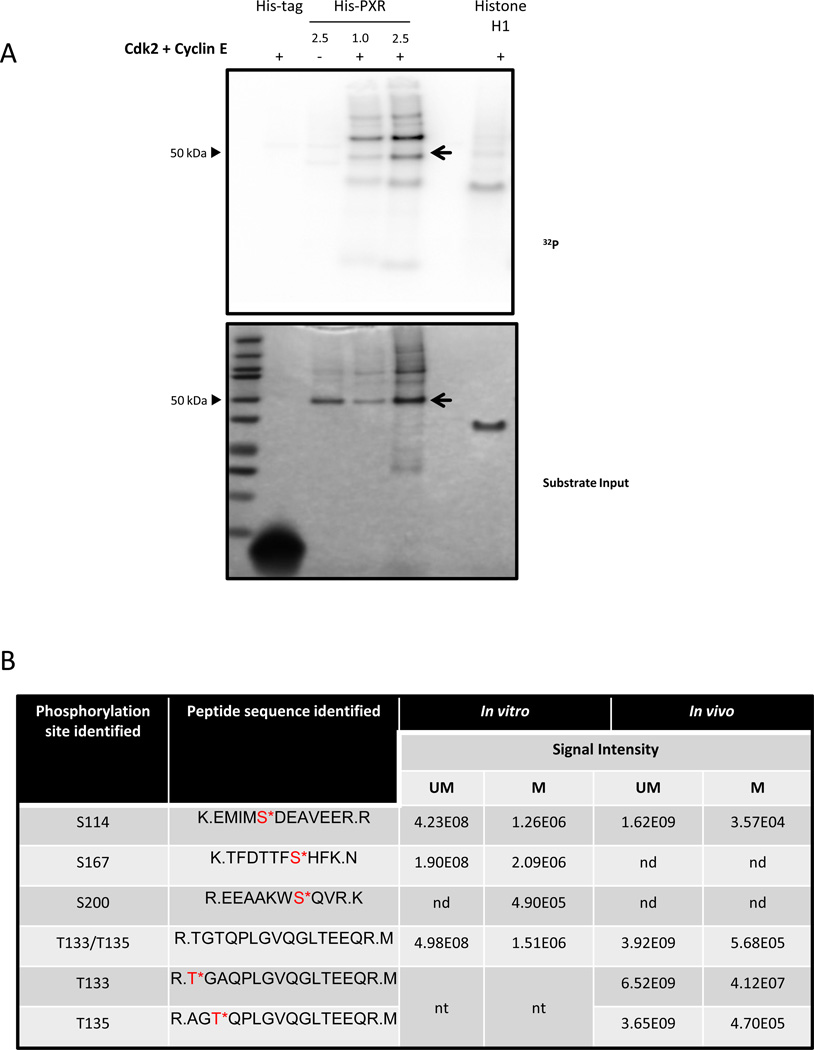
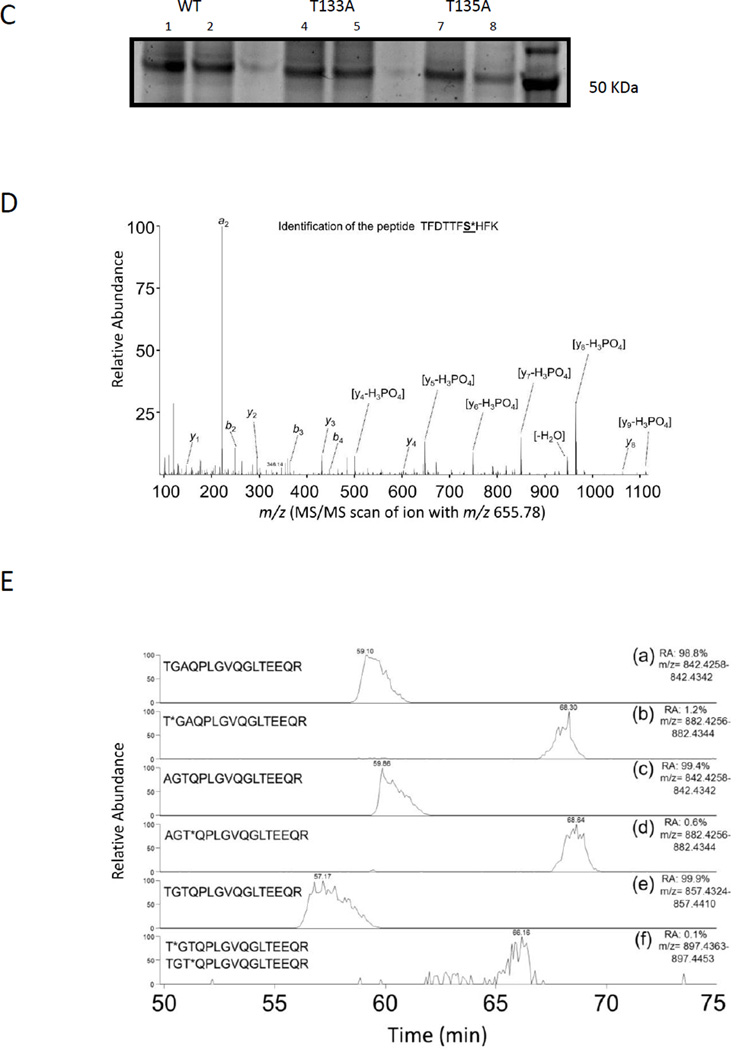
Figure 1. PXR is phosphorylated in vitro and in cells.
(A) His-PXR (1 or 2.5 µg) was incubated at 37°C for 30 min with Cdk2 and cyclin E along with [γ-32P]-ATP. Samples were resolved on a 4–12% gradient gel, and [γ-32P]-ATP incorporation was visualized using a phosphor screen (upper panel), and protein amounts in the samples were detected by SimplyBlue staining of the gel (lower panel). Histone H1 and His-tag were used as a positive and negative substrate control, respectively. The PXR band was indicated with an arrow. (B) Phosphorylation sites identified by using mass spectrometry analysis in His-PXR WT phosphorylated by Cdk2/cyclin E in vitro, and in Flag-PXR WT, Flag-PXR T133A, or Flag-PXR T135A immunoprecipitated from HEK293T cells transiently transfected with corresponding plasmid (in vivo). Serine or threonine residues followed by an asterisk (*) indicate phosphorylated residues; UM = unmodified peptide; M = phosphorylated peptide; nd = not detected; nt = not tested. Signal intensities are calculated from area under the curve for the detected precursor ions. (C) Anti-Flag immunoprecipitated samples prepared from HEK293T cells transiently overexpressing either Flag-PXR WT (lanes 1 & 2) or mutants Flag-PXR T133A (lanes 4 & 5) or Flag-PXR T135A (lanes 7 & 8) were resolved on gradient gel and stained using Sypro Ruby stain. (D) Modified peptide sequence TFDTTFS*HFK (asterisk indicating serine phosphorylation), was identified based on assignment of multiple product ions (b and y ions) in the MS/MS scan of the precursor ion at M/z 665.78. The phosphorylation of serine 167 was confirmed based on the assignment of characteristic “y-H3PO4” ions and other ions (based on a mass loss of 97.9769 Da). (E) Extracted-ion chromatography (XIC) of wild type and mutant PXR sequences showing elution times and signal intensities for the non-modified peptide as well as the singly phosphorylated peptide. Panel (a) and (b) are derived from the immunoprecipitated T133A sample and show the TGAQPLGVQGLTEEQR and T*GAQPLGVQGLTEEQR, respectively. Panel (c) and (d) are derived from the immunoprecipitated T135A sample and show the AGTQPLGVQGLTEEQR and AGT*QPLGVQGLTEEQR, respectively. Panel (e) and (f) are derived from the immunoprecipitated PXR WT sample and show the TGTQPLGVQGLTEEQR and T*GTQPLGVQGLTEEQR/ TGT*QPLGVQGLTEEQR, respectively. Relative abundance (RA) of the signals of the corresponding peptides is noted for each XIC.