Abstract
The biochemical regulatory network downstream of receptor tyrosine kinase (RTK) signaling is controlled by two opposing ETS family members: the transcriptional activator Pointed (Pnt) and the transcriptional repressor Yan. A bistable switch model has been invoked to explain how pathway activation can drive differentiation by shifting the system from a high-Yan/low-Pnt activity state to a low-Yan/high-Pnt activity state. Although the model explains yan and pnt loss-of-function phenotypes in several different cell types, how Yan and Pointed protein expression dynamics contribute to these and other developmental transitions remains poorly understood. Toward this goal we have used a functional GFP-tagged Pnt transgene (Pnt-GFP) to perform a comparative study of Yan and Pnt protein expression throughout Drosophila development. Consistent with the prevailing model of the Pnt-Yan network, we found numerous instances where Pnt-GFP and Yan adopt a mutually exclusive pattern of expression. However we also observed many examples of co-expression. While some co-expression occurred in cells where RTK signaling is presumed low, other co-expression occurred in cells with high RTK signaling. The instances of co-expressed Yan and Pnt-GFP in tissues with high RTK signaling cannot be explained by the current model, and thus they provide important contexts for future investigation of how context-specific differences in RTK signaling, network topology, or responsiveness to other signaling inputs, affect the transcriptional response.
Keywords: Drosophila, ETS transcription factor, Embryo, Imaginal discs, EGFR signaling, Gene regulation
Introduction
Signaling downstream of Receptor Tyrosine Kinases (RTKs) regulates cell fate specification, collective cell migration, and cell proliferation during development (Sopko and Perrimon, 2013). In Drosophila, RTK signaling converges on the Ras/MAPK cascade, which regulates a transcriptional response controlled by two ETS transcription factors, the activator Pointed (Pnt) and the repressor Yan (O’Neill et al., 1994; Rebay and Rubin, 1995). pnt encodes two isoforms, pntP1 and pntP2, in which distinct 5′ exons are spliced into common 3′ exons to create two proteins with similar DNA binding activity but different amino-terminal activation domains (Klambt, 1993; Scholz et al., 1993). As a result, PntP1 is a constitutively active transcription factor while PntP2 activity requires stimulation by RTK signaling (Gabay et al., 1996; O’Neill et al., 1994).
The current model of Yan-Pnt interactions is as follows. Prior to RTK signaling, the intrinsically weaker DNA binding affinity of Pnt proteins relative to Yan ensures that Yan prevails to repress transcriptional targets (O’Neill et al., 1994; Rebay and Rubin, 1995; Xu et al., 2000). Upon RTK signaling, activated MAPK phosphorylates Yan, triggering its nuclear export and degradation (O’Neill et al., 1994; Rebay and Rubin, 1995). Concomitant MAPK-mediated phosphorylation of PntP2 converts it into a strong activator, which then turns on transcription of targets previously repressed by Yan (Brunner et al., 1994; O’Neill et al., 1994). PntP1 is itself an RTK target gene whose induction is thought to ensure sustained activation of gene expression after the initial transient RTK signal has decayed (Shwartz et al., 2013). Mathematical modeling of this network has described a bistable system in which cells transition between two states: an OFF state in which high Yan and low Pnt activity maintain a progenitor-like cell state; and an ON state in which high Pnt and low Yan activity promote cell fate specification by activating appropriate target gene expression (Graham et al., 2010).
Although the computational model correctly predicts cell fate specification defects that occur in certain yan or pnt mutants (Graham et al., 2010), achieving molecular-level understanding of Yan-Pnt behavior during these transitions has been hindered by the lack of suitable antibody reagents to document the dynamics of Pnt protein expression. Thus, while in situ hybridization and reporter-based analyses have hinted at complex pnt expression dynamics (Klambt, 1993; Morimoto et al., 1996; Scholz et al., 1993; Shwartz et al., 2013), the low cellular resolution of the former and the potential problems with reporter protein perdurance in the latter have made accurate side-by-side comparisons of Yan versus Pnt expression difficult.
Here we present a high-resolution atlas that compares the expression patterns of Yan and Pnt proteins across multiple stages of oogenesis, embryogenesis and imaginal development, with particular attention to contexts where RTK signaling is known to be important. To do this we used a recombineered Pnt genomic transgene that was generated in a collaboration with the ModEncode project. GFP was inserted in-frame at the carboxy-terminus, tagging both P1 and P2 isoforms. Pnt-GFP rescues pnt null mutants to viable and fertile adults, indicating full function of the transgene. Our results have revealed that in most tissues where RTK signaling occurs, the expression patterns of Pnt-GFP and Yan are mutually exclusive, with only transient co-expression, as predicted by the bistable state model. However, several notable exceptions to this rule were found in which Pnt-GFP and Yan were co-expressed in cells with active RTK signaling. In tissues where RTK signaling is low, we also observed extensive co-expression of Pnt-GFP and Yan. Together our work both supports many of the basic tenets of the current Yan-Pnt interaction model and challenges several of its underlying assumptions.
Material and methods
Drosophila genetics
Pnt-GFP was recombineered using the ModEncode pipeline and inserted into the VK0037 landing site; details are available through FlyBase and flies can be obtained from the Bloomington Stock Center. Pnt-GFP rescued the following amorphic heteroallelic combinations to full viability and fertility: pntΔ88/pnt2 and pntΔ88/Df(pnt)Exel9012. Larval and pupal tissues were dissected from w1118; Pnt-GFP; pntΔ88/pnt2 animals while ovaries and embryos were obtained from w1118; Pnt-GFP animals. No qualitative differences in Pnt-GFP expression were noted between these genetic backgrounds.
Immunohistochemistry
Embryos were dechorionated in 50% bleach and fixed in 1:1 heptane/4% formaldehyde for 30 minutes at room temperature. Ovaries were fixed in 3:1 heptane/4% formaldehyde with 0.025% NP40 for 30 minutes at room temperature and then permeabilized in 1% Triton X-100 for 30 minutes. Imaginal tissues were fixed for 30 minutes in 4% paraformaldehyde. Primary and secondary antibodies were incubated either for 2 hours at room temperature or overnight at 4°C.
Primary antibodies used were: rabbit anti-GFP (1:2000, Molecular Probes); mouse anti-Eve 3C10 (1:10, Developmental Studies Hybridoma Bank (DSHB)); mouse anti-Yan 8B12H9 (1:100 ovaries, 1:500 embryos, 1:200 imaginal discs and brain, DSHB); mouse anti-β-galactosidase 40-1a (1:50, DSHB); mouse anti-β-galactosidase (1:1000, Promega); guinea-pig anti-Yan (1:10,000, (Webber et al., 2013)); rat anti-Elav 7E8A10 (1:1000, DSHB); rabbit anti-Atonal (1:2500, Jarman et al,1995). Secondary antibodies used were: DyLight goat anti-mouse Cy3, goat anti-rabbit 488 and goat anti-guinea pig Cy5 (1:2000, Jackson Immuno Research); goat anti-mouse, goat anti-rat and goat anti-rabbit Alexa Fluor 488, Alexa 546, Alexa 633, Pacific Blue (1:200, Invitrogen). 72mM DAPI was used to mark nuclei. Tissues were mounted in VectaShield (Vector Laboratories) or ProLong Antifade (Life Technologies). Images of egg chambers, embryos and pupal discs were acquired on Zeiss LSM 510 confocals. Larval tissues were imaged on a Leica Sp5 confocal. Optical sections were projected in Volocity 5.5 (Perkin Elmer) and the Leica LAS AF Image Lite and Zeiss LSM Image browsers.
Results and Discussion
A Fluorescent Reporter for Pnt
To visualize Pnt protein expression, we recombineered GFP coding sequence into the pnt open reading frame such that both the Pnt-P1 and Pnt-P2 protein products would have GFP fused to their carboxy termini. This recombineering was performed on a bacterial artificial chromosome (BAC) that contained 90.7 kb of the Drosophila genome, including the pnt gene. The modified DNA was inserted into a P[acman] transformation vector (Venken et al., 2006), and inserted into the Drosophila genome by ΦC31-mediated integration. Flies carrying one or two copies of Pnt-GFP in either a wildtype or pnt−/− background were equally viable and fertile, and patterns of Pnt-GFP expression appeared unaffected by overall locus copy number. We conclude that the Pnt-GFP transgene can functionally replace both isoforms of pnt, and that the GFP tag has no deleterious effect on Pnt protein functions. We then used this novel reagent to survey Pnt and Yan protein expression dynamics across many stages of development, from oogenesis to pupal imaginal discs.
Egg chamber development
RTK signaling is required at multiple stages of Drosophila egg chamber development to specify posterior and dorsal fates, and to direct border cell migration (reviewed in (Horne-Badovinac and Bilder, 2005). Expression patterns and loss of function phenotypes have implicated yan and pnt in these events, although their interactions are not clear from the published data. For example, loss of yan delays border cell migration (Schober et al., 2005), while loss of pnt compromises dorsal follicle cell fates, leading to the formation of a single dorsal appendage (Boisclair Lachance et al., 2009; Morimoto et al., 1996; Zartman et al., 2009). Our comparison of Yan and Pnt-GFP expression revealed both overlapping and complementary patterns, with more complex dynamics than previously appreciated (Price and Lai, 1999; Schober et al., 2005)(Morimoto et al., 1996).
Pnt-GFP was initially detected throughout the germarium and germline, but rapidly became enriched in the posterior such that by stage 6 it was detected exclusively in posterior follicle cells of the egg chamber (Fig. 1A, B). In contrast, Yan was strongly expressed in the stalk cells that connect the egg chambers, and in all follicle cells, with highest expression at the termini by stage 6 (Fig. 1A, B). This initial co-expression of Yan and Pnt-GFP in posterior follicle cells coincides with the stage when EGFR signaling breaks the symmetry along the AP axis by inducing posterior follicle cell fate (González-Reyes et al., 1995; Roth et al., 1995). Given that MAPK activation is involved (Dammai and Hsu, 2003; Ghiglione et al., 1999; Zartman et al., 2011), we predicted rapid downregulation of posterior Yan protein. However, downregulation was not complete until stage 8 (Fig. 1C), well after the initial signaling event. Thus, either Yan is not a MAPK target during specification of posterior fate, or its downregulation by MAPK is not rapid. One possibility is that Yan expression is maintained via Notch signaling, which is required to pattern posterior follicle cell fates (González-Reyes and St Johnston, 1998) and has been shown to activate yan expression in other contexts (Rohrbaugh et al., 2002; Schober et al., 2005). However, increased yan transcription cannot on its own explain the co-expression of Yan and Pnt-GFP in the presence of active MAPK, given that overexpressed Yan protein is effectively cleared in tissues where RTK signaling is active (Rebay and Rubin, 1995), even when ectopic levels are five times that of the endogenous protein (Melen et al., 2005).
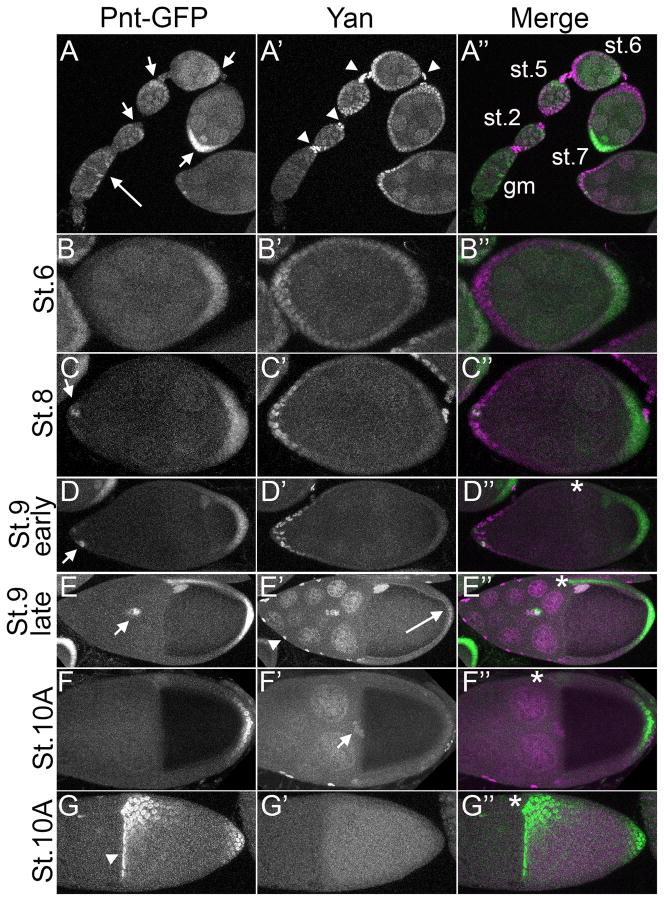
Figure 1. Pnt-GFP expression overlaps with Yan expression in early egg chambers and during border cell migration.
All images are lateral projections of the relevant optical slices except G–G″ which represent a dorsal projection of an egg chamber. Anterior is left and where relevant, dorsal is up and marked by an asterisk. For the merge, Pnt-GFP is in green and Yan in magenta. A–A″: Ovariole showing germarium (gm) to stage 7 egg chambers. Pnt-GFP expression is detected in the gm (long arrow), while Yan expression is below detectable levels. Starting in stage 2 egg chambers, Pnt-GFP is most prominent in posterior follicle cells (FCs), (short arrows). Yan is more uniformly expressed in all FCs, with highest expression of Yan detected in stalk cells (arrowheads). Note that Pnt-GFP appears to be both nuclear and cytoplasmic. B–B″: By stage 6, Pnt-GFP expression is restricted to posterior FCs while Yan expression is maintained at high levels in anterior FCs and to a lower extent posterior FCs in contact with the oocyte. C–C″: Yan expression is present in anterior FCs in stage 8 egg chambers. Pnt-GFP is mostly restricted to posterior FCs, but a few GFP-positive FCs are detected at the anterior tip (arrow). D–D″: In early stage 9 egg chambers, anterior expression of Pnt-GFP has increased (arrow). Pnt-GFP in dorso-posterior FCs appears as a gradient. Weak Pnt-GFP expression is visible in the oocyte nucleus. E–E″: Migrating border cells (BCs) express both Pnt-GFP and Yan (arrow) at late stage 9, as does the oocyte nucleus. Yan is highly expressed in stretch cells covering the nurse cells (arrowheads) and in nurse cell nuclei. Weak expression is detected in posterior FCs (long arrow). High Pnt-GFP expression is visible at the dorsal anterior and posterior region of the follicular epithelium, while intermediate levels of expression are visible in-between these regions. F–F″: By stage 10A, once BCs have finished their migration towards the oocyte, Yan expression is maintained but Pnt-GFP is lost (arrow). High expression of Pnt-GFP is maintained in posterior FCs. Yan expression appears to have dropped off everywhere except the stretch cells (arrowhead) G–G″: Dorsal view of stage 10A egg chamber showing strong Pnt-GFP expression in the dorsal midline region and in the anteriormost follicle cells of the dorsal region (arrowhead). Yan expression has fallen below detection threshold.
Co-expression of Yan and Pnt-GFP was also detected in the border cells prior to and during migration (Fig. 1D–E). Once the border cells reached the nurse cell-oocyte border, expression of Pnt-GFP was lost while Yan levels decayed more slowly (Fig. 1F). As with the posterior follicle cells, persistence of Yan protein is somewhat incongruous given RTK signaling is active in migrating border cells (Dammai and Hsu, 2003; Zartman et al., 2011), and so would be expected to target Yan for rapid degradation. Rapid loss of Pnt expression was also not predicted from the standard bistable switch model. Thus either Yan and/or Pnt are not serving as RTK/MAPK effectors in this context, or the regulatory interactions are quite different from those predicted by the bistable model.
The third patterning event requiring RTK signaling is the establishment of dorsal-ventral (DV) oocyte polarity at stage 9 (reviewed in (Horne-Badovinac and Bilder, 2005). Both Yan and Pnt-GFP expression were detected in the oocyte nucleus at this stage, but only Pnt-GFP was detected in the dorsal follicle cells (Fig. 1D, E). By stage 10A, a DV gradient of Pnt-GFP expression had developed (Fig. 1E, G), consistent with the role of pnt in patterning dorsal fates (Boisclair Lachance et al., 2009; Morimoto et al., 1996; Zartman et al., 2009). The lack of Yan expression in the dorsal follicle cells suggests Yan-independent functions for Pnt in patterning dorsal fates.
Embryogenesis
Consistent with the numerous roles played by RTK signaling during embryogenesis, yan and pnt mutants exhibit complex phenotypes, with defects in ventral midline patterning (Gabay et al., 1996; Melen et al., 2005), specification of mesodermally derived fates (Halfon et al., 2000), tracheal morphogenesis (Caviglia and Luschnig, 2013; Hacohen, 1997; Klambt, 1993; Myat et al., 2005; Ohshiro et al., 2002; Samakovlis et al., 1996) and cephalogenesis (Jurgens et al., 1984; Rogge et al., 1995). As described below, the patterns of Yan and Pnt-GFP expression reveal extensive complementarity in regions of active MAPK signaling and extensive overlap elsewhere.
The first RTK signaling event occurs at the termini of the embryo where the receptor Torso is activated and promotes specification of head and tail fates. Although Torso acts through the Ras/MAPK cascade, the downstream transcriptional response does not seem to require either Pnt or Yan (Sopko and Perrimon, 2013). Consistent with this, we did not detect expression of either protein prior to stage 4 (not shown).
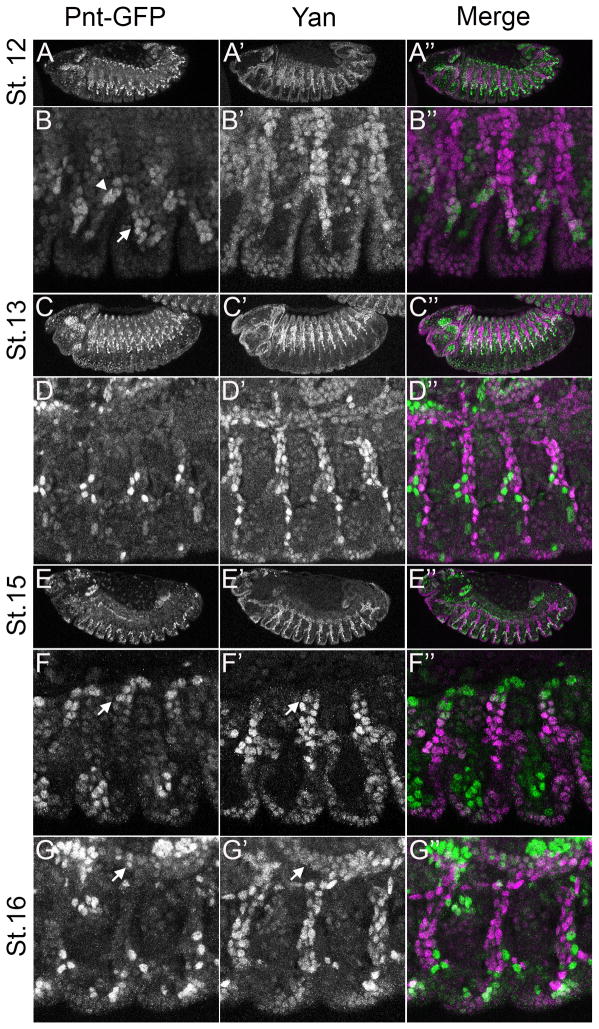
As reported previously (Price and Lai, 1999), Yan expression was first detected in the pole cells at stage 4 (Fig. 2A) and then in all cells of the stage 5 cellular blastoderm (Fig. 2B). Pnt-GFP was first detected at stage 6 in an anterior cap of cells from which Yan had been markedly downregulated (Fig. 2C). Yan levels were also reduced at the posterior region, but remained high in the pole cells. Throughout the trunk of the embryo, Yan adopted a weakly striped pattern reminiscent of pair-rule gene expression. Since active MAPK is reported to be low at stage 6 (Gabay et al., 1997a), the Yan stripes may reflect transcriptional regulation by the gap genes. By stage 7, Pnt-GFP expression appeared at the posterior region, setting up a predominantly complementary pattern with Yan that was maintained through stage 9 (Fig. 2D–E). These mutually exclusive patterns of Pnt-GFP and Yan are consistent with known domains of MAPK activation at these stages (Cinnamon et al., 2008; Gabay et al., 1997a, Gabay et al., 1997b), with the most striking example around the ventral midline (Fig. 2E–G).
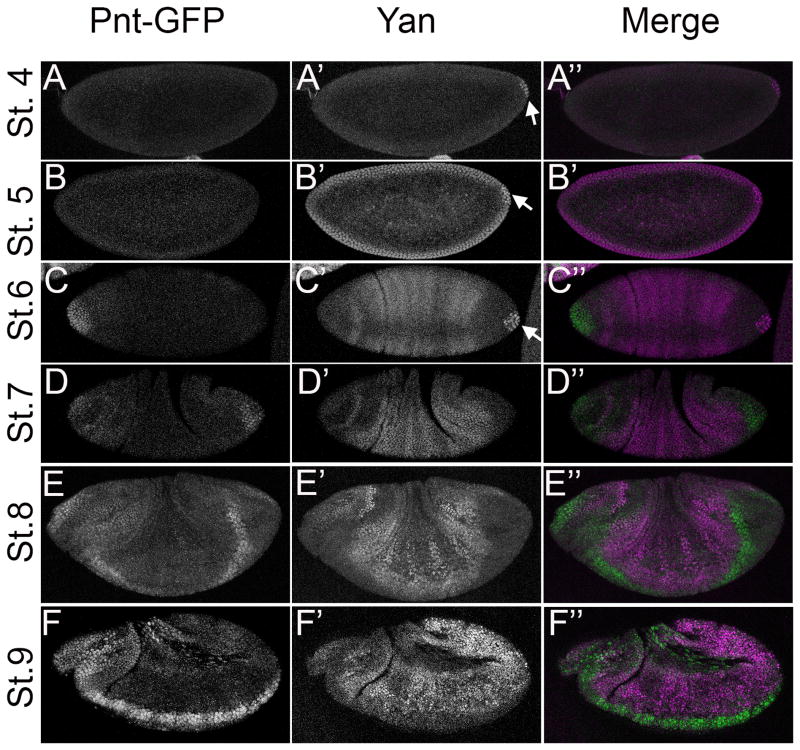
Figure 2. Pnt-GFP and Yan have mostly non-overlapping expression patterns in the early embryo.
All images are projections of the relevant optical slices (except G–G″) of embryos carrying two copies of Pnt-GFP. Lateral view of early to mid-stage embryos (except C–C″ which show a dorsal view) are oriented anterior to the left and dorsal up. For the merge, Pnt-GFP is in green and Yan in magenta. A–A″: Yan expression is detected first in pole cells at posterior of stage 4 embryos (arrow). B–B″: Yan is expressed after cellularization in all cells of the blastoderm and in pole cells (arrow). C–C″: Pnt-GFP expression is first detected in an anterior cap of cells at stage 6. Yan expression remains strong in pole cells (arrow) but is lost in the somatic cells at both termini. Moderate expression is detected in a pattern of lateral stripes, with reduced expression near the dorsal midline. D–D″: By stage 7, Pnt-GFP expression is detected at both termini, while Yan expression increases in lateral regions. E–E″: Expression is essentially complementary at stage 8, with Pnt-GFP in ventral cells. Low levels of lateral Pnt-GFP expression overlap with Yan. F–F″: At stage 9, Pnt-GFP expression is strong in the region of the developing brain and ventral nerve cord and weaker everywhere else except for the amnioserosa (arrow). Yan expression appears in a complementary pattern.
The mutually exclusive pattern of Yan and Pnt-GFP expression around the ventral midline represents one of the classic examples around which the bistable switch model was developed (Graham et al., 2010; Melen et al., 2005; Rebay and Rubin, 1995). Indeed, in the 3–4 rows of cells around the midline where MAPK is active (Gabay et al., 1997a), only Pnt-GFP was present (Fig. 3A–B). Yan remained excluded from this high Pnt-GFP expression domain, except in segmentally arrayed clusters of unidentified cells (Fig. 3D′ arrow). More laterally, cells of the ventral ectoderm co-expressed Pnt-GFP and Yan (Fig 3C–D). As the cells invaginated to form the mesoderm, the stripe of Pnt-GFP expressing cells around the midline narrowed, but with continued exclusion of Yan (Fig. 3E–F).
Figure 3. Complementary Pnt-GFP and Yan expression in the ventral neuroectoderm.
All images are ventral projections of embryos carrying two copies of Pnt-GFP and oriented anterior is to the left. The ventral midline (M) is indicated by an arrow. For the merge, Pnt-GFP is in green and Yan in magenta. A–A″: Pnt-GFP expression is enriched ventrally where Yan is excluded. B–B″: Close-up view of the embryo in A–A″ showing the ventral domain of high Pnt-GFP flanked by a lateral domain of high Yan. Pnt-GFP is strongly expressed in 3–4 rows of cells on each side of the midline (M) and weakly expressed beyond. Yan is not detectable in any of these rows but is expressed at higher levels than Pnt in ventral-lateral cells. C–C″: By stage 11, Pnt-GFP has increased throughout the embryo, but the highest levels remain in the ventral neuroectoderm where Yan expression is absent. D–D″: Close-up view of the stage 11 embryo in C–C′ highlights three domains of differential Pnt-GFP/Yan expression: 2–3 rows of ventral-most cells each side of the midline have high Pnt-GFP and mostly lack Yan; ventro-lateral cells express high Yan and low Pnt-GFP; and more lateral cells have moderate levels of Pnt-GFP and low Yan. A segmentally repeating pattern of small clusters of cells expressing both high Yan and Pnt-GFP is seen in the lateral and intermediate rows (arrows). E–E″: At stage 12, only the lateral row of the ventral Pnt-GFP expression remains. Yan expression is absent from these lateral rows, and strong in immediately flanking cells, which also have moderate Pnt-GFP expression. F–F″: Higher magnification view of the embryo in E–E″.
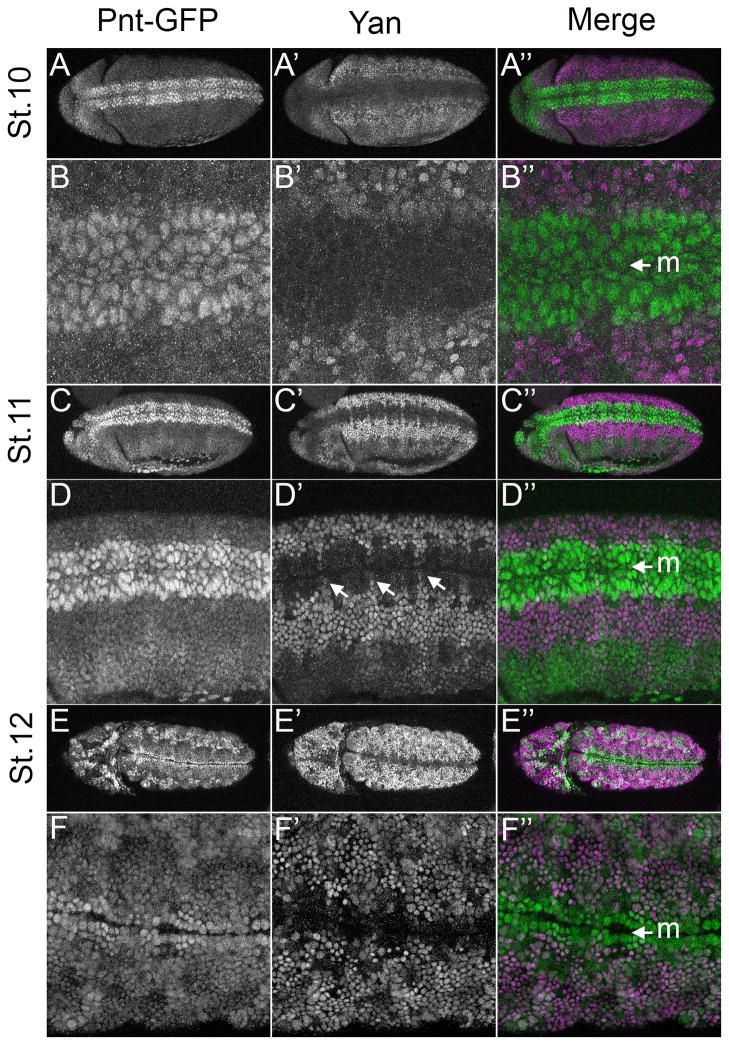
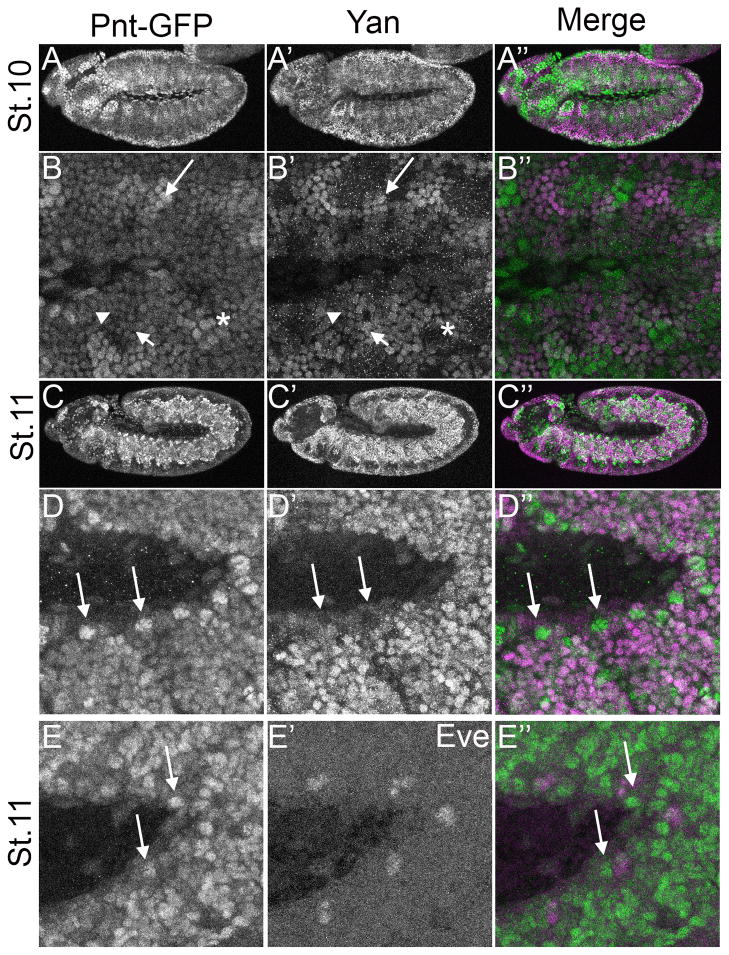
The patterning and morphogenesis of the trachea is another context in which RTK signaling acts through pnt and yan (reviewed in (Affolter et al., 2003; Amin Ghabrial et al., 2003; Hogan and Kolodziej, 2002), and as predicted, the two display a complementary pattern of expression. The trachea arises from the fusion of segmentally repeated tracheal trees whose branching patterns depend on the RTK Breathless (Btl), a member of FGFR family. Btl activates MAPK in the tip cells (TC) at the ends of each of the 6 primary branches, which in turn targets Yan and Pnt to regulate secondary branch formation. Thus, loss of pnt results in the loss of secondary branches (Klambt, 1993; Myat et al., 2005; Samakovlis et al., 1996) while loss of yan induces excessive secondary branching (Caviglia and Luschnig, 2013; Hacohen, 1997; Ohshiro et al., 2002). Consistent with these phenotypes, Yan was expressed throughout the trachea except at the TCs (Caviglia and Luschnig, 2013; Ohshiro et al., 2002), while Pnt-GFP was strongest in the TCs of all six branches (Fig. 5A). Despite their largely complementary expression patterns (Fig. 4A–G), Pnt-GFP and Yan overlapped in 2–3 cells adjacent to the TCs of the lateral anterior and posterior ganglion tracheal branches at stage 12 (Fig. 5B arrowhead and arrow respectively), in migrating cells of the dorsal branch (Fig. 4E–F) and in dorsal trunk cells after migration was complete (Fig. 4G).
Figure 5. Coexpression of Pnt-GFP and Yan in the developing mesoderm.
All images are lateral projections of embryos carrying two copies of Pnt-GFP. For the merge, Pnt-GFP is in green and Yan in magenta. A–A″: With the exception of the amnioserosa and some head structures, stage 10 embryos exhibit extensively overlapping Pnt-GFP and Yan expression. B–B″: Higher magnification of the embryo depicted in A–A″ shows that although most cells express both Yan and Pnt-GFP, their levels are not uniform: some cells express low levels of both proteins (arrowhead); some express high levels of one and low levels of the other (short arrow); some show high levels of both (long arrow); and a few cells have high Pnt-GFP but no Yan (asterisk). C–C″: Both Pnt-GFP and Yan expression appear upregulated in the mesoderm at stage 11. D–D″: High magnification view of the embryo in C–C″ highlights the extensive overlap, but also identifies a segmentally arrayed cluster of cells that only express high Pnt-GFP (long arrows). E–F″: These cell clusters do not overlap with Eve expression in the heart muscle precursors at stage 11 and onwards (not shown).
Figure 4. Complementarity of Pnt-GFP and Yan expression during tracheal development.
All images are lateral projections of embryos carrying two copies of Pnt-GFP and oriented anterior is to the left and dorsal is up. For the merge, Pnt-GFP is in green and Yan in magenta. A–B″: Pnt-GFP and Yan expression overlap in lateral anterior trunk branches (arrowhead) and in lateral posterior branches (arrow). C–C″: Most tracheal cells at this stage express high Yan levels whereas strong Pnt-GFP is only seen in 3–4 cells per segment. D–D″: Close-up view of tracheal branches imaged in C–C″. The Pnt-GFP expressing cells flank a region of high Yan expressing cells (arrow). E–E″: At stage 15, most tracheal cells continue to express high Yan and low Pnt-GFP. F–F″: Some overlap is visible in the dorsal branch in the zoomed in view of the stage 15 embryo (arrow). G–G″: High magnification of a stage 16 embryo shows that both Pnt-GFP and Yan are expressed in the dorsal trunk (arrow).
In contrast to the complementary patterns observed around the ventral midline and the trachea, extensive Yan/Pnt-GFP overlap occurred in the mesoderm, with peak expression at stage 11 (Fig. 5A–D). Possibly the low levels of activated MAPK in this tissue (Gabay et al., 1997a, Gabay et al., 1997b; Liu et al., 2006) permit stable accumulation of Yan. A few instances of complementary expression were observed, most noticeably in segmentally arrayed dorsal cell clusters that expressed high Pnt-GFP but not Yan (Fig. 5D). These clusters were adjacent to the pericardial cells expressing Even-skipped (Eve), whose specification requires RTK signaling through Yan and Pnt (Fig. 5E) (Halfon et al., 2000). Given their position relative to the Eve-positive clusters, these cells are likely cardiac cells that co-express Lde and the Tbx-20 transcription factors Midline and H15 (Qian et al., 2005). Yan was also prominently expressed throughout the epidermis, where only very low Pnt-GFP was detected (Fig. 5C).
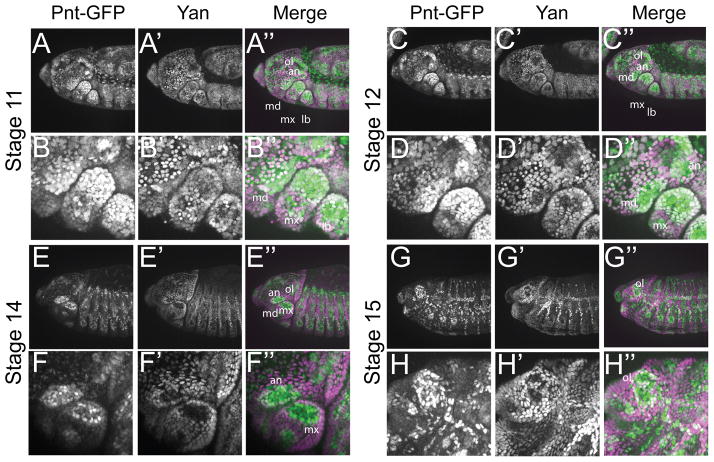
Patterning of the embryonic head involves RTK signaling (Chang et al., 2003; Dumstrei et al., 2002, 1998). yan mutants display an anterior open phenotype that has been suggested to result from overproliferation of the dorsal neuroectoderm and a failure of the epidermal layer covering the brain to migrate over the involuting lobes (Jurgens et al., 1984; Rogge et al., 1995). In contrast, pnt mutants have reduced head structures that result in a pointed head skeleton (Jurgens et al, 1984). Prior to head involution, Pnt-GFP was detected in both the procephalon and in the mandible, maxillary and labial segments in a pattern complementary to that of Yan (data not shown). At stages 11 and 12, most cells within the head had either low Yan and high Pnt-GFP, or the converse (Fig. 6A and C). However, coincident Yan and Pnt-GFP occurred in cells within the gnathal segments and the antennal and optic lobes (Fig. 6B and D) at a stage when active RTK signaling is occurring (Dumstrei et al., 1998; Dumstrei et al., 2002). By stage 14, expression of Yan and Pnt-GFP resolved into complementary patterns, with co-expression only detected in cells surrounding the presumptive antennal and optic lobe (Fig. 6E–H). The co-expression of Yan and Pnt-GFP in the mandible segment was intriguing given the role of this segment in the formation of the larval mouth hooks, which are missing in the yan mutant (Rogge et al., 1995). Like the antennal and optic lobe, the mandible segment also exhibited high levels of active MAPK staining (Chang et al., 2003; Dumstrei et al., 2002, 1998).
Figure 6. Yan and Pnt-GFP expression during head formation.
All images are lateral projections of embryos carrying two copies of Pnt-GFP and oriented anterior is to the left and dorsal is up. For the merge, Pnt-GFP is in green and Yan in magenta. A–A″: At stage 11, Pnt-GFP and Yan levels increase in the head region. Low level of overlap is seen throughout the head. Strong overlap is visible in the optic lobe (ol) and the antennal lobe (an) as well as the in the mandillar (md), maxillary (mx) and labial gnathal segments. B–B″: Close-up view of the head shown in B–B″. C–C″: At stage 12, the overlap between Pnt-GFP and Yan remains strong in the ol and the an, but begins to decrease in the md, mx and lb segments. D–D″: Close-up view of head structures shown in D–D″. E–E″: The overlap between Pnt-GFP and Yan remains strong only in the ol. In the an, Pnt-GFP and Yan begin to adopt a complementary pattern. A mutually exclusive pattern of expression between Pnt-GFP and Yan is visible in gnathal segments. F–F″: Close-up view of an and mx regions shown in F–F″. G–G″: By stage 15, Pnt-GFP and Yan adopt a complementary pattern in most of the head. At the center of the ol region, Yan expression is gone while Pnt-GFP expression remains strong. Strong Pnt-GFP and Yan co-expression is observable around this central region. H–H″: Closeup view of the head structures shown in G–G″.
Larval Wing, Leg and Antennal Imaginal Discs
Post-embryonic pattern formation in Drosophila occurs primarily in the larval imaginal discs that are the anlagen of adult body structures. Appendages such as wings, legs and antennae require patterning not only across the DV and AP axes, but also along the proximal-distal (PD) axis. RTK signaling contributes extensively to patterning these axes. For example, EGFR signaling and pnt are required for PD axis specification in the leg, and presumably the antenna, where patterning mechanisms are quite similar (Estella et al., 2012; Galindo et al., 2005). In the third instar wing, activated MAPK is detected around the presumptive wing margin and vein regions (Gabay et al., 1997b; Martín-Blanco et al., 1999; Reich et al., 1999), and this pattern parallels the expression of the protease rhomboid (rho), which contributes to EGFR ligand activation (Guichard et al., 1999; Sturtevant et al., 1993).
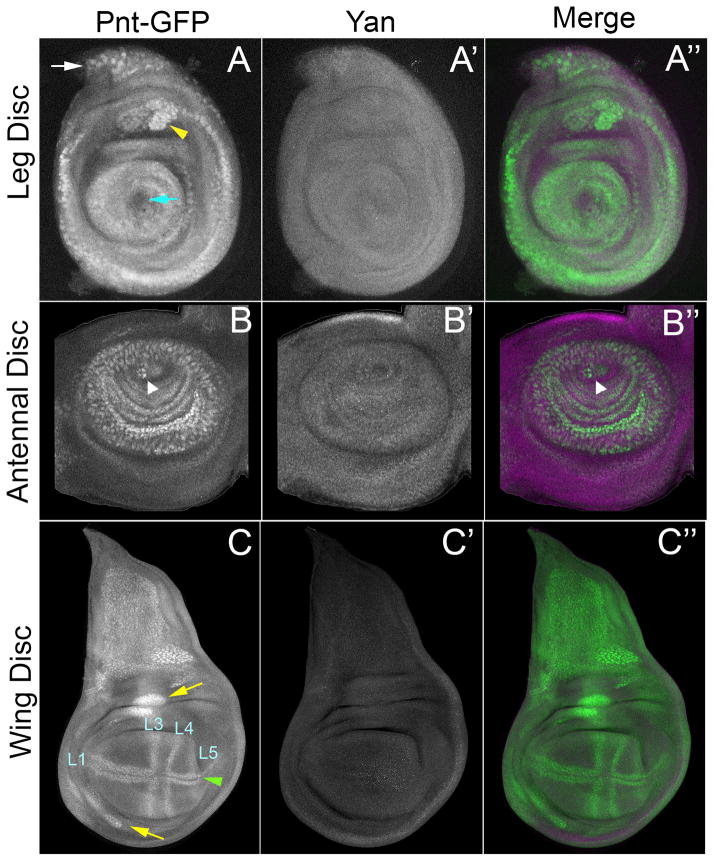
However, unlike most other developmental contexts in which Yan-Pnt interactions determine EGFR output, only Pnt-GFP was expressed in these three larval imaginal discs. Yan levels were below our detection threshold. The Pnt-GFP patterns overlapped with known domains of activated MAPK (Fig. 7A–C). In the leg (Fig. 7A), Pnt-GFP expression occurred in most leg segment primordia and in a region of the pleura (white arrow). It was absent in the central region that gives rise to the claws (blue arrow). A cluster in the dorsal-most femur segment (yellow arrowhead) possibly gives rise to the edge bristle. In the antenna (Fig. 7B), Pnt-GFP was detected in a series of concentric rings corresponding to sensory organ precursors of the A2 and A3 anlagen; these were located around the presumptive arista (white arrowhead). In the wing (Fig. 7C), Pnt-GFP was expressed in a subdomain of the notum, in clusters in the dorsal and ventral hinge region (yellow arrows), and in a cross-like pattern of stripes in the wing pouch. The pair of stripes flanking the AP boundary corresponded to the presumptive L3 and L4 wing veins. The stripes flanking the DV boundary were more complex. In the anterior compartment, these stripes were thicker and likely corresponded to proneural regions that form the L1 vein. In the posterior compartment, the stripes overlapped with presumptive sensory organ precursors that form posterior margin bristles (green arrowhead) and with the presumptive L5 vein. This expression pattern in the wing pouch is similar to expression of rho, which activates EGFR signaling (Sturtevant et al., 1993). However, the presumptive L2 vein region expresses rho but did not express detectable Pnt-GFP, suggesting that other transcription effectors might mediate EGFR signaling in that region.
Figure 7. Pnt-GFP and Yan expression in third instar imaginal discs.
All images are confocal projections. (A–A″) Leg disc. (B–B″) Antennal disc. (C–C″) Wing disc. Pnt-GFP (A, B, C), Yan (A′, B′, C′), and a merge of the two (A″, B″, C″) are shown. The merge shows Pnt-GFP (green) and Yan (purple) in the same image. For the wing disc, presumptive Pnt-GFP-positive wing veins are indicated. All images are oriented anterior-left and dorsal-up.
The absence of Yan in these imaginal discs raises the question of what alternate mechanisms are used to ensure precision to Pnt-mediated regulation of target gene expression in response to RTK signaling. One possibility is that another transcription factor might provide MAPK-responsive repression analogous to that normally conferred by Yan. An intriguing candidate is Capicua (Cic), which acts downstream of RTK signaling in multiple developmental contexts, and whose transcriptional repressor activity, like that of Yan, is attenuated by MAPK phosphorylation (Roch et al., 2002) (reviewed in (Jiménez et al., 2012)). Given the similarities between Yan and Cic, it is tempting to speculate that Pnt and Cic might be found in bistable interactions in tissues where Yan expression is not detected. Alternatively, perhaps the other signaling pathways such as Notch, Dpp and Wg that contribute to these patterning events, restrict the competence of cells to respond to RTK signaling, therefore limiting the ability of Pnt to induce target gene expression in a Yan-independent manner.
Pupal wing
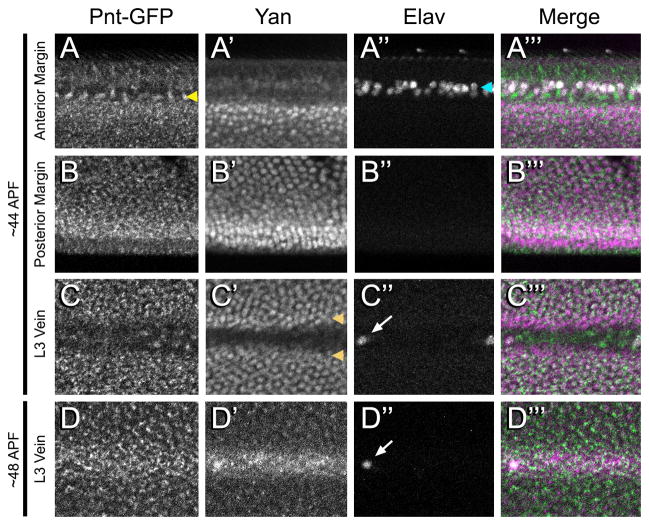
The requirement for EGFR signaling to pattern the vein/intervein regions extends beyond the third larval instar (reviewed in (Blair, 2007)), prompting us to examine the expression of Pnt-GFP and Yan during pupal stages. The wing disc everts during early pupal development such that the dorsal and ventral halves of the pouch become overlaid and stretched along the proximal-distal axis to form the wing blade proper. Subsequent to eversion, we observed dynamic patterns of Yan and Pnt-GFP expression. In contrast to the late larval disc where Yan was undetectable, approximately 44h after puparium formation (APF), Yan was detected in nuclei of inter-vein cells throughout the wing (Fig. 8A–C). Pnt-GFP was present in the same inter-vein cells as Yan, but was localized exclusively to the cytoplasm (Fig. 8A–C). Pnt-GFP was also present in a row of cells adjacent to the anterior Elav-positive bristle neurons (Fig. 8A) and at low levels in cells within the veins (Fig. 8C–C‴). Yan was not detected in these cells, and Pnt-GFP subcellular localization appeared more nuclear than in the intervein region (Fig. 8A″). The opposite pattern was observed in a row of cells at each vein/inter-vein border (Fig 8C). Thus all combinations of Yan/PntGFP colocalization and mutual exclusion were observed, suggesting significant complexity to their interactions.
Figure 8. Pnt-GFP and Yan expression in the pupa wing.
A–Ctprime;: A wing from a pupa ~44h APF showing a region from the anterior wing margin (A–A‴), posterior wing margin (B–B‴), and L3 wing vein (C–C‴). Note the adjacent rows of Pnt-GFP cells (yellow arrowhead) and Elav cells (cyan arrowhead) along the anterior margin. Also note the two rows of cells bordering the L3 vein (orange arrowheads) that exclusively contain Yan. D–D‴: A wing from a pupa ~48h APF showing the L3 wing vein. Pnt-GFP (A–D), Yan (A′–D′), Elav (A″–D″) and a merge (A‴–D‴) are shown. The merge shows Pnt-GFP (green), Yan (purple) and Elav (white) in the same image. The region of the L3 vein shown in C and D is mid-wing, where the left-most Elav-positive cell is the L3.1 sensory cell (arrows). All images are oriented anterior-up and distal-left.
The expression of Yan and Pnt-GFP evolved further such that at 48hr APF, expression of both was reduced in the intervein cells and increased in the vein cells (Fig 8D). The reduction in inter-vein cell expression was particularly striking for Yan. This shift from inter-vein to vein expression matches the known dynamics of activated MAPK and pntP1 mRNA expression (Guichard et al., 1999; Martín-Blanco et al., 1999; Wessells et al., 1999).
Two unanticipated aspects of these expression patterns warrant further comment. First, the cytoplasmic localization of Pnt-GFP was unexpected, as cytoplasmic functions for Pnt have not been described. Second, Pnt-GFP and Yan appear co-expressed, albeit in different subcellular compartments, at times and places when EGFR signaling and MAPK is active. This suggests there must be mechanisms to protect Yan from MAPK phosphorylation or to block the nuclear export that normally ensues. Perhaps analogous to mechanisms used in the early embryo (Kim et al., 2011; Kim et al., 2010), substrate competition could limit the efficiency with which MAPK phosphorylates Yan. Cic provides a potential candidate competitor based on its expression patterns and EGFR signaling responsiveness in this tissue (Roch et al., 2002). Yan could provide the reciprocal service for Cic, allowing the two repressors to collaboratively mediate appropriate transcriptional repression. Such redundancy could explain why yan loss-of-function phenotypes have not been reported in the wing, despite its dynamic pupal expression pattern.
Visual development
Patterning of the third instar larval eye disc is coordinated by a morphogenetic furrow (MF) that moves across the disc from posterior to anterior. A complex network of signals acts in the MF to organize the first wave of cells to commit to a terminal fate: the R8 photoreceptors. One key factor in this process is the bHLH transcription factor Atonal (Ato) (Jarman et al., 1994). Ato is initially expressed in a stripe of cells immediately anterior to the MF. As the MF passes, these cells restrict their Ato expression into alternating clusters of expressing and non-expressing cells. The clusters of Ato-expressing cells are known as Intermediate Groups (IGs) (Rawlins et al., 2003). Cells within each IG signal to one another via lateral inhibition to further restrict Ato expression to one cell, which becomes the R8 photoreceptor (Lee et al., 1996). Subsequent cell fate specification occurs in a stereotyped sequence of events that each requires activation of EGFR signaling by R8-derived signals. Each R8 cell secretes an EGFR ligand which activates signaling in neighboring cells to induce the following sequence of cell fates: R2/R5 – R3/R4 – R1/R6 – R7 – Cone cells (reviewed in (Tsachaki and Sprecher, 2012)). The resulting clusters of 8 photoreceptors and 4 cone cells constitute the nascent ommatidium or unit eye.
Both Pnt-GFP and Yan were detected in the MF and regions posterior to the MF (Fig. 9A). Pnt-GFP was first detected in the stripe of Ato-positive undifferentiated (precursor) cells immediately anterior to the MF (Fig. 9B, C, green arrow); Yan was not detected in these cells. The evolution of the Ato pattern closely correlated with changes in Pnt-GFP and Yan expression. As the Ato stripe became restricted, cells began to produce Yan protein and further increased their expression of Pnt-GFP (Fig. 9C, cyan arrow). By the time that Ato was restricted to IGs, Yan and Pnt-GFP were becoming differentially expressed. The Ato-positive cells of the IGs expressed high levels of Pnt-GFP but little or no Yan (Fig. 9C, white arrowhead). Conversely, Ato-negative cells in between the IGs expressed Yan as well as Pnt-GFP (Fig. 9C, orange arrow). This trend continued as Ato expression resolved to the R8 cell, which was positive for Pnt-GFP and negative for Yan (Fig 9C, white arrow).
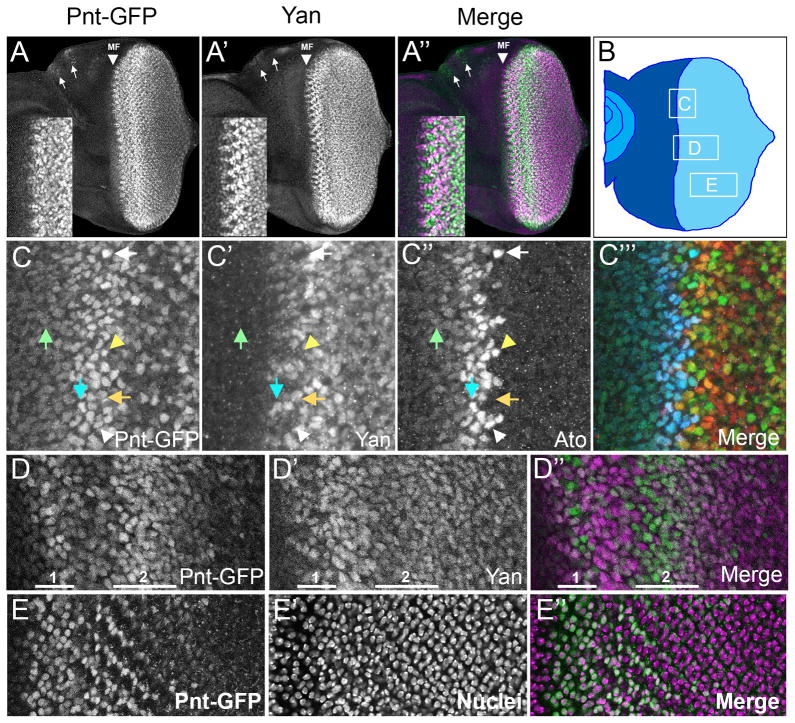
Figure 9. Pnt-GFP and Yan expression in the larval eye imaginal disc.
(A–A″) Confocal projection of Pnt-GFP (green) and Yan (purple) in the developing eye disc. Position of the morphogenetic furrow (MF) is indicated, as are the ocelli (arrows). Insets show a magnified view of Yan and Pnt-GFP near the MF. (B) Diagram of an eye imaginal disc with the MF bisecting the undifferentiated anterior region (dark blue) and differentiating posterior region (light blue). Boxes outline the approximate locations of regions magnified in the subsequent panels of this figure. Letters inside boxes correspond to specific panels. (C–C‴) Expression of Pnt-GFP (C), Yan (C′), and Ato (C″) in a zone that spans the MF and includes cells at different stages of Ato expression, arrayed from anterior to posterior. The first stage has a broad band of anterior cells co-expressing low levels of Pnt-GFP and Ato (green arrow). At the onset of the second stage, Ato and Pnt-GFP expression increases, and some cells also begin to express Yan (cyan arrow). This stage has intermediate groups (IGs) expressing high levels of Ato and Pnt-GFP but not expressing Yan (white arrowhead). The cells between each IG express no Ato, but do express high levels of Pnt-GFP and increased levels of Yan (orange arrow). Two or three posterior cells in each IG express the highest level of Ato, and these cells are the R8 equivalence group (yellow arrowhead). The third stage has a single Ato-positive cell, which is fated to become the R8 cell (white arrow). Pnt-GFP levels remain high or even increase in this cell, while Yan is not detected above background. The merge (C‴) shows Pnt-GFP (green), Yan (red) and Ato (blue) in the same image. (D–D″) Expression of Pnt-GFP (green) and Yan (purple) in uncommitted precursor cells posterior to the MF. Highlighted 1 and 2 are two successive regions of induction of Pnt-GFP (these do not correspond to the stages of Ato expression described in C). Note that Yan is broadly expressed in precursors and is upregulated in the zone spanning the two regions of Pnt-GFP induction. More posterior, the precursor cells lose Pnt-GFP. (E–E″) Expression of Pnt-GFP (green) in cells within the second wave of induction (left) and in differentiating photoreceptors (right). The protein is primarily nuclear (purple). All images are oriented anterior-left and dorsal-up.
Immediately posterior to the stripe of high Pnt-GFP expression in the MF (Fig. 9D highlighted as 1), Pnt-GFP expression was reduced, giving the appearance of a posterior stripe of high Yan and low Pnt-GFP. This Pnt-GFP reduction was transient, and was followed by a broader stripe of strong Pnt-GFP expression (Fig. 9D highlighted as 2). This wave was longer lasting, with a decay of Pnt-GFP towards the posterior margin. Yan expression in and posterior to zone 2 was more uniform in undifferentiated cells, with a less pronounced decay towards the posterior.
It was also possible to follow Yan and Pnt-GFP expression in cells that were committing to differentiated fates. As had been previously observed (Price and Lai, 1999; Rebay and Rubin, 1995), Yan expression dropped in differentiating photoreceptor cells (Fig. 10A–C). Unexpectedly, Pnt-GFP was present in the nuclei of the differentiating photoreceptors but its expression was not maintained over time (Fig. 9E). Thus, older photoreceptors located in progressively more posterior positions had reduced and ultimately undetectable Pnt-GFP expression (Fig. 10A–C, inset 10B, arrows). The kinetics of loss of Pnt-GFP expression was rapid such that Pnt-GFP expression coincided only transiently with expression of Elav, one of the earliest protein expression markers for photoreceptor neuron differentiation (Hart et al., 1990) (Fig. 10E). The reduction of Pnt-GFP correlated with a shift in its subcellular localization from nucleus to cytoplasm before completely decaying (Fig. 10F). A different pattern was seen in the non-neuronal cone cells where Pnt-GFP and Yan proteins were co-expressed, with a slower decay compared to that observed in the photoreceptors (Fig. 10D). To summarize, our data indicate that photoreceptor differentiation correlates with a parallel decay of both Yan and Pnt-GFP proteins, although the decay in Pnt-GFP is preceded by a burst of Pnt-GFP expression as cells commit to a terminal fate.
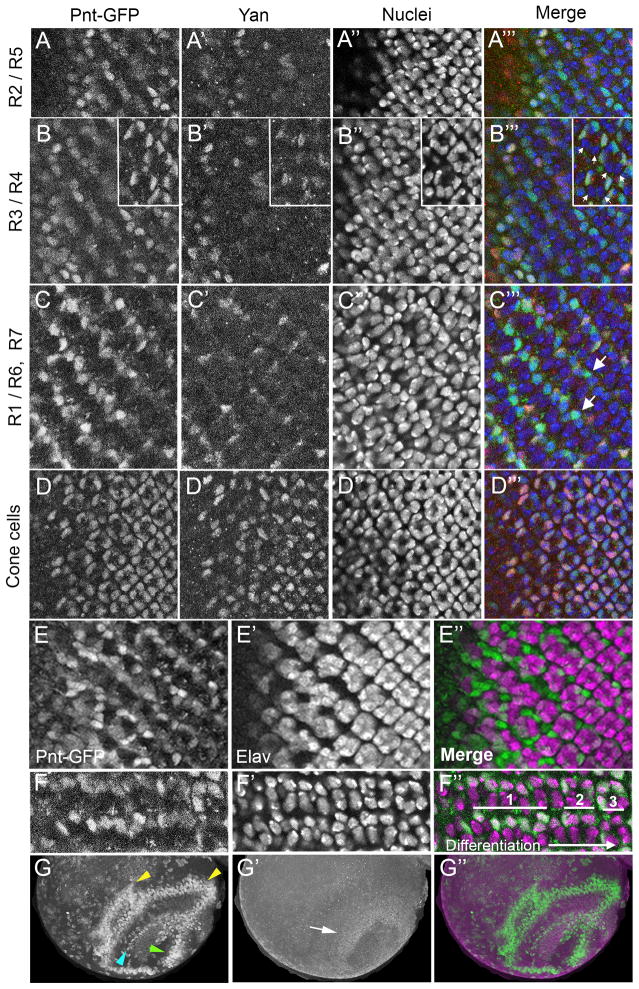
Figure 10. Pnt-GFP and Yan expression in the visual system.
All images are oriented anterior-left and dorsal-up. (A–A‴) Ommatidia located immediately posterior to the morphogenetic furrow, soon after R2/R5 commitment. Shown is a section containing R2 and R5 cell nuclei. (B–B‴) Sections through ommatidia highlighting R3 and R4 cell nuclei. Ommatidia in the main panel are at the stage soon after R3/R4 commitment. The inset shows ommatidia at the two-cone-cell stage, many hours after R3/R4 commitment. Arrows mark R3 and R4 nuclei, which display a lack of Yan and Pnt-GFP. (C–C‴) A section through ommatidia highlighting R1, R6, and R7 cell nuclei. Anterior ommatidia (left) are at a stage shortly after R1/R6 commitment. These ommatidia show Pnt-GFP expression in R1 and R6 nuclei that decreases as the ommatidia mature – those ommatidia more posterior (right). Arrows highlight R7 nuclei that are Pnt-GFP positive. This expression is also transient. (D–D‴) Ommatidia that are at various stages of cone cell commitment. The section shows cone cell nuclei, which co-express Pnt-GFP and Yan. Pnt-GFP (A–D), Yan (A′–D′), DAPI-stained nuclei (A″–D″), and the merge (A‴–D‴) are shown. The merge is colored for Pnt-GFP (green), Yan (red), and nuclei (blue). (E–E″) A section through a disc where the left side shows ommatidia with cells undergoing photoreceptor fate commitment and the right side shows ommatidia at stages after fate commitment (right). The section displays differentiating photoreceptor nuclei, which express the Elav protein (purple). These nuclei generally lack Pnt-GFP (green). Pnt-GFP instead is prominent in the nuclei of cells undergoing fate commitment and not yet expressing Elav. (F–F″) Highly magnified view of Pnt-GFP (green) in two rows of ommatidia from a section showing photoreceptor nuclei (purple). Their differentiation progresses from left to right. Photoreceptors highlighted with 1 have Pnt-GFP primarily in the nucleus, while photoreceptors highlighted with 2 have primarily cytoplasmic Pnt-GFP. Photoreceptors highlighted with 3 show reduction of cytoplasmic Pnt-GFP. (G–G″) Dorsal view of one optic lobe in the third instar brain showing Pnt-GFP (green) and Yan (purple). Stripes of Pnt-GFP (yellow and cyan arrowheads) in the medulla and a cluster of Pnt-GFP (green arrowhead) in the lamina. These are complementary to a ring of Yan-positive cells (white arrowhead).
Photoreceptors project their axons to the optic lobes of the brain, and in so doing, they trigger the progressive differentiation of interneurons from neuroepithelial cells. EGFR signals travel in a wave across the neuroepithelium as part of the differentiation process (Yasugi et al., 2010). As expected, Pnt-GFP was expressed in a stripe of cells corresponding to the proneural wave (Fig. 10G, cyan arrowhead). Other stripes of Pnt-GFP were detected in the medulla region of differentiating cells (yellow arrowheads), and a region of Pnt-GFP expression in the differentiating lamina (green arrowhead) likely corresponding to the reported Pnt-P1 expression in laminar glia (Yasugi et al., 2010). Yan was enriched in a ring of undifferentiated neuroepithelial cells that separate the lamina and medulla (Fig. 10G′ white arrow), where Pointed is expressed only at low levels. Overall, the expression of Yan and Pnt-GFP in the optic lobes was mostly complementary (Fig. 10G″).
The expression of Pnt and Yan is largely unknown at later stages of visual development, which progress throughout pupation. We investigated Pnt-GFP/Yan expression in the eye at a mid-pupal stage. At this stage, every eye cell’s nucleus has a defined position within the pseudostratified epithelium. For example, the apical region of the eye contains the cone cell nuclei and cytoplasmic projections from the other cells. Yan protein remained abundant within the cone cell nuclei whereas Pnt-GFP was not detected (Fig. 11A). However, Pnt-GFP appeared in the apical cytoplasm of the photoreceptor cells. Pnt-GFP was also detected in the perinuclear cytoplasm and nuclei of photoreceptor cells, albeit at lower levels (Fig. 11B). Yan was also abundant in primary pigment cells (Fig. 11B) and slightly less abundant in the secondary and tertiary pigment cells (Fig. 11C). Collectively these results show mutually exclusive expression of Pnt-GFP and Yan in the terminally differentiated non-neuronal cell types of the mid-pupal eye. Moreover, Pnt-GFP was detected primarily in the cytoplasm. Whether this localization results from the GFP tag preventing efficient degradation of Pnt or whether Pnt has cytoplasmic signaling functions remain open questions.
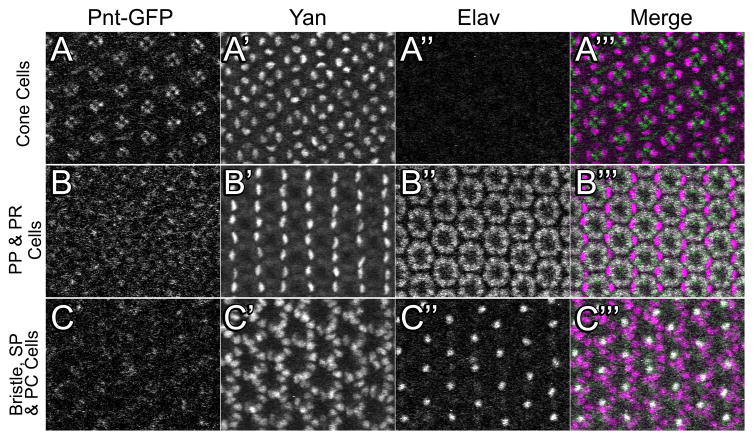
Figure 11. Pnt-GFP and Yan expression in the 48hr APF pupal eye.
Three optical sections from the same eye are shown with 25 ommatidia in the field of view. For each section, Pnt-GFP (A, B, C), Yan (A′, B′, C′), Elav (A″, B″, C″), and a merge (A‴, B‴, C‴) are shown. The merge shows Pnt-GFP (green), Yan (purple) and Elav (white) in the same image. (A–A‴) An apical section showing the cytoplasmic projections of photoreceptor (PR) cells and the nuclei of the cone cells. (B–B‴) A medial section showing primary pigment (PP) cell nuclei and Elav-positive PR cell nuclei. (C–C‴) A basal section showing interommatidial bristle cell nuclei (bristle neurons are positive for Elav), secondary pigment (SP) cell nuclei, and tertiary pigment (TP) cell nuclei. All images are oriented anterior-left and dorsal-up.
Concluding Remarks
Our analysis of the expression patterns of Pnt-GFP and Yan during development has revealed that both proteins exhibit a more complex relationship than previously appreciated. As predicted by the standard bistable model, the expression of Pnt-GFP and Yan are mutually exclusive in many tissues where RTK signaling occurs. However, an important finding from our analysis is the extensive co-expression of Pnt-GFP and Yan in many tissues at diverse stages of development. We have observed co-expression in posterior follicle cells and during border cell migration in early oogenesis, in embryonic mesoderm and head structures, in cells surrounding the intermediate groups and in cone cells during eye development, and in intervein cells during pupal wing development. These tissues should provide tractable contexts in which to explore how the balance between upstream signaling inputs that regulate yan and pnt transcription and the downstream post-transcriptional and post-translational mechanisms that regulate translation and protein localization and stability are fine-tuned to achieve stable co-expression.
While co-expression of Pnt-GFP and Yan appears to contradict the standard model, it does not exclude a bistable model in which factor activity, rather than expression, switches as a result of RTK signaling. For example, MAPK phosphorylation of these factors might alter their abilities to recognize target genes or regulate transcription of these genes. In this scenario, Yan phosphorylation would reduce its potency as a transcriptional repressor, enabling Pnt to activate its targets. Whether or not this interpretation is correct, a mechanism would have to be in place to protect Yan from MAPK-mediated cytoplasmic translocation and degradation. One potential mechanism for the presence of Yan in a context where RTK is active would be the presence of substrate competition between the different MAPK targets (Kim et al., 2011, Kim et al., 2010), which would prevent the interaction of activated MAPK with Yan. Another mechanism could be association of Yan with specific proteins that protect Yan from phosphorylation leading to degradation. Finally, Yan might be differentially sensitive to MAPK phosphorylation, such that phosphorylation of some but not all sites is not sufficient to trigger degradation. Conditions where MAPK phosphorylates all sites on Yan might be required for degradation. In this regard, it is worth noting that the retinal cone cells and embryonic mesoderm receive modest EGFR signaling that translates to modest MAPK activation (Gabay et al., 1997a, 1997b; Kumar et al., 2003; Liu et al., 2006). The retinal R7 cell also receives the modest EGFR signal, but it additionally activates the Sevenless RTK, which boosts MAPK activation in this cell. The R7 cell shows a more rapid reduction in Yan, consistent with the notion that degradation could require more extensive Yan phosphorylation. It is important to note that eventually in many of the tissues where Yan and Pnt-GFP overlap, the co-expression pattern eventually resolves to a more mutually exclusive pattern, which might reflect a change in the type of decision RTK signaling controls. Alternatively, a switch in expression kinetics might occur from the onset of the RTK signal, but the kinetics are slow enough that there is a delay before a steady state pattern is achieved.
In conclusion, our observations of the patterns of expression of Pnt-GFP and Yan have revealed greater complexity to the transcriptional networks downstream of RTK signaling than previously appreciated. While the standard bistable switch model correctly predicts expression patterns of Yan and Pnt-GFP in most contexts, co-expression of Pnt-GFP and Yan in the presence of activated MAPK suggests either differential sensitivity to MAPK, alternate network topology, responsiveness to alternate signaling events, or a combination of the above.
Acknowledgments
Yuh Nung Jan generously gave us anti-Atonal antibody. We are grateful to Kevin White and the Recombineering Core (U. Chicago), Thom Kauffmann (U. Indiana), and the Biological Imaging Facility (Northwestern University). N.P. was funded by the Chicago Biomedical Consortium and an International Student Research Fellowship from the Howard Hughes Medical Institute (HHMI). J.L.W. was supported by an award from the American Heart Association. This work was supported by National Institutes of Health grant P50GM081892. Further support from R01GM077581 (R.W.C) and R01 GM80372 (I.R.) are acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Affolter M, Itoh N, Shilo B, Thiery J, Werb Z. Tube or Not Tube : Remodeling Epithelial Tissues by Branching Morphogenesis Branching morphogenesis involves the restructuring. Development. 2003;4:11–18. doi: 10.1016/s1534-5807(02)00410-0. [DOI] [PubMed] [Google Scholar]
- Amin Ghabrial SL, Metzstein Mark M, Krasnow MA. BRANCHING MORPHOGENESIS OF THE DROSOPHILA TRACHEAL SYSTEM. 2003 doi: 10.1146/annurev.cellbio.19.031403.160043. [DOI] [PubMed] [Google Scholar]
- Blair SS. Wing vein patterning in Drosophila and the analysis of intercellular signaling. Annual review of cell and developmental biology. 2007;23:293–319. doi: 10.1146/annurev.cellbio.23.090506.123606. [DOI] [PubMed] [Google Scholar]
- Boisclair Lachance JF, Fregoso Lomas M, Eleiche A, Bouchard Kerr P, Nilson LA. Graded Egfr activity patterns the Drosophila eggshell independently of autocrine feedback. Development Cambridge England. 2009;136:2893–2902. doi: 10.1242/dev.036103. [DOI] [PubMed] [Google Scholar]
- Brunner D, Dücker K, Oellers N, Hafen E, Scholz H, Klämbt C. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature. 1994;370:386–9. doi: 10.1038/370386a0. [DOI] [PubMed] [Google Scholar]
- Caviglia S, Luschnig S. The ETS domain transcriptional repressor Anterior open inhibits MAP kinase and Wingless signaling to couple tracheal cell fate with branch identity. Development (Cambridge, England) 2013;140:1240–9. doi: 10.1242/dev.087874. [DOI] [PubMed] [Google Scholar]
- Chang T, Shy D, Hartenstein V. Antagonistic relationship between Dpp and EGFR signaling in Drosophila head patterning. Developmental biology. 2003;263:103–13. doi: 10.1016/s0012-1606(03)00448-2. [DOI] [PubMed] [Google Scholar]
- Cinnamon E, Helman A, Ben-Haroush Schyr R, Orian A, Jiménez G, Paroush Z. Multiple RTK pathways downregulate Groucho-mediated repression in Drosophila embryogenesis. Development (Cambridge, England) 2008;135:829–37. doi: 10.1242/dev.015206. [DOI] [PubMed] [Google Scholar]
- Dammai V, Hsu T. EGF-dependent and independent activation of MAP kinase during Drosophila oogenesis. The anatomical record Part A Discoveries in molecular cellular and evolutionary biology. 2003;272:377–382. doi: 10.1002/ar.a.10037. [DOI] [PubMed] [Google Scholar]
- Dumstrei K, Nassif C, Abboud G, Aryai A, Hartenstein V. EGFR signaling is required for the differentiation and maintenance of neural progenitors along the dorsal midline of the Drosophila embryonic head. Development (Cambridge, England) 1998;125:3417–26. doi: 10.1242/dev.125.17.3417. [DOI] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Shy D, Tepass U, Hartenstein V. Interaction between EGFR signaling and DE-cadherin during nervous system morphogenesis. Development (Cambridge, England) 2002;129:3983–94. doi: 10.1242/dev.129.17.3983. [DOI] [PubMed] [Google Scholar]
- Estella C, Voutev R, Mann RS. A dynamic network of morphogens and transcription factors patterns the fly leg. Current topics in developmental biology. 2012;98:173–98. doi: 10.1016/B978-0-12-386499-4.00007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay L, Scholz H, Golembo M, Klaes A, Shilo B, Klambt C. EGF receptor signaling induces pointed P1 transcription and inactivates Yan protein in the Drosophila embryonic ventral ectoderm. Development. 1996;122:3355–3362. doi: 10.1242/dev.122.11.3355. [DOI] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ. MAP kinase in situ activation atlas during Drosophila embryogenesis. Development (Cambridge, England) 1997a;124:3535–41. doi: 10.1242/dev.124.18.3535. [DOI] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ. In situ activation pattern of Drosophila EGF receptor pathway during development. Science (New York, NY) 1997b;277:1103–6. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- Galindo MI, Bishop SA, Couso JP. Dynamic EGFR-Ras signalling in Drosophila leg development. Developmental dynamics : an official publication of the American Association of Anatomists. 2005;233:1496–508. doi: 10.1002/dvdy.20452. [DOI] [PubMed] [Google Scholar]
- Ghiglione C, Carraway KL, Amundadottir LT, Boswell RE, Perrimon N, Duffy JB. The transmembrane molecule kekkon 1 acts in a feedback loop to negatively regulate the activity of the Drosophila EGF receptor during oogenesis. Cell. 1999;96:847–856. doi: 10.1016/s0092-8674(00)80594-2. [DOI] [PubMed] [Google Scholar]
- González-Reyes A, Elliott H, St Johnston D. Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature. 1995;375:654–8. doi: 10.1038/375654a0. [DOI] [PubMed] [Google Scholar]
- González-Reyes A, St Johnston D. Patterning of the follicle cell epithelium along the anterior-posterior axis during Drosophila oogenesis. Development Cambridge England. 1998;125:2837–2846. doi: 10.1242/dev.125.15.2837. [DOI] [PubMed] [Google Scholar]
- Graham TGW, Tabei SMA, Dinner AR, Rebay I. Modeling bistable cell-fate choices in the Drosophila eye: qualitative and quantitative perspectives. Development (Cambridge, England) 2010;137:2265–78. doi: 10.1242/dev.044826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard A, Biehs B, Sturtevant MA, Wickline L, Chacko J, Howard K, Bier E. rhomboid and Star interact synergistically to promote EGFR/MAPK signaling during Drosophila wing vein development. Development (Cambridge, England) 1999;126:2663–2676. doi: 10.1242/dev.126.12.2663. [DOI] [PubMed] [Google Scholar]
- Hacohen N. Branching morphogenesis in the Drosophila tracheal system. Stanford University; Stanford, California: 1997. [Google Scholar]
- Halfon MS, Carmena A, Gisselbrecht S, Sackerson CM, Jiménez F, Baylies MK, Michelson AM. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103:63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- Hart AC, Krämer H, Van Vactor DL, Paidhungat M, Zipursky SL. Induction of cell fate in the Drosophila retina: the bride of sevenless protein is predicted to contain a large extracellular domain and seven transmembrane segments. Genes & Development. 1990;4:1835–1847. doi: 10.1101/gad.4.11.1835. [DOI] [PubMed] [Google Scholar]
- Hogan BLM, Kolodziej PA. Organogenesis: molecular mechanisms of tubulogenesis. Nature Reviews Genetics. 2002;3:513–523. doi: 10.1038/nrg840. [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S, Bilder D. Mass transit: epithelial morphogenesis in the Drosophila egg chamber. Developmental dynamics : an official publication of the American Association of Anatomists. 2005;232:559–74. doi: 10.1002/dvdy.20286. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. Atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- Jiménez G, Shvartsman SY, Paroush Z. The Capicua repressor--a general sensor of RTK signaling in development and disease. Journal of cell science. 2012;125:1383–91. doi: 10.1242/jcs.092965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G, Wieschaus E, Nusslein-Volhard C, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. II. Zygotic loci on the third chromosome. Rouxs Arch Dev Biol. 1984;193:283–295. doi: 10.1007/BF00848157. [DOI] [PubMed] [Google Scholar]
- Kim Y, Andreu MJ, Lim B, Chung K, Terayama M, Jiménez G, Berg CA, Lu H, Shvartsman SY. Gene regulation by MAPK substrate competition. Developmental cell. 2011;20:880–7. doi: 10.1016/j.devcel.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Coppey M, Grossman R, Ajuria L, Jiménez G, Paroush Z, Shvartsman SY. MAPK substrate competition integrates patterning signals in the Drosophila embryo. Current biology : CB. 2010;20:446–51. doi: 10.1016/j.cub.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klambt C. The Drosophila gene pointed encodes two ETS-like proteins which are involved in the development of the midline glial cells. Development. 1993;117:163–176. doi: 10.1242/dev.117.1.163. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Hsiung F, Powers MA, Moses K. Nuclear translocation of activated MAP kinase is developmentally regulated in the developing Drosophila eye. Development (Cambridge, England) 2003;130:3703–14. doi: 10.1242/dev.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Hu X, Yu S, Baker N. The scabrous gene encodes a secreted glycoprotein dimer and regulates proneural development in Drosophila eyes. Mol Cell Biol. 1996;16:1179–1188. doi: 10.1128/mcb.16.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Qian L, Wessells RJ, Bidet Y, Jagla K, Bodmer R. Hedgehog and RAS pathways cooperate in the anterior-posterior specification and positioning of cardiac progenitor cells. Developmental biology. 2006;290:373–85. doi: 10.1016/j.ydbio.2005.11.033. [DOI] [PubMed] [Google Scholar]
- Martín-Blanco E, Roch F, Noll E, Baonza A, Duffy JB, Perrimon N. A temporal switch in DER signaling controls the specification and differentiation of veins and interveins in the Drosophila wing. Development (Cambridge, England) 1999;126:5739–47. doi: 10.1242/dev.126.24.5739. [DOI] [PubMed] [Google Scholar]
- Melen GJ, Levy S, Barkai N, Shilo BZ. Threshold responses to morphogen gradients by zero-order ultrasensitivity. Molecular systems biology. 2005;1:0028. doi: 10.1038/msb4100036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto AM, Jordan KC, Tietze K, Britton JS, O’Neill EM, Ruohola-Baker H. Pointed, an ETS domain transcription factor, negatively regulates the EGF receptor pathway in Drosophila oogenesis. Development Cambridge England. 1996;122:3745–3754. doi: 10.1242/dev.122.12.3745. [DOI] [PubMed] [Google Scholar]
- Myat MM, Lightfoot H, Wang P, Andrew DJ. A molecular link between FGF and Dpp signaling in branch-specific migration of the Drosophila trachea. Developmental biology. 2005;281:38–52. doi: 10.1016/j.ydbio.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Ohshiro T, Emori Y, Saigo K. Ligand-dependent activation of breathless FGF receptor gene in Drosophila developing trachea. Mechanisms of Development. 2002;114:3–11. doi: 10.1016/s0925-4773(02)00042-4. [DOI] [PubMed] [Google Scholar]
- Price MD, Lai Z. The yan gene is highly conserved in Drosophila and its expression suggests a complex role throughout development. Dev Genes Evol. 1999;209:207–17. doi: 10.1007/s004270050245. [DOI] [PubMed] [Google Scholar]
- Qian L, Liu J, Bodmer R. Neuromancer Tbx20-related genes (H15/midline) promote cell fate specification and morphogenesis of the Drosophila heart. Developmental biology. 2005;279:509–24. doi: 10.1016/j.ydbio.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Rawlins EL, White NM, Jarman AP. Echinoid limits R8 photoreceptor specification by inhibiting inappropriate EGF receptor signalling within R8 equivalence groups. Development (Cambridge, England) 2003;130:3715–24. doi: 10.1242/dev.00602. [DOI] [PubMed] [Google Scholar]
- Rebay I, Rubin GM. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell. 1995;81:857–66. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- Reich A, Sapir A, Shilo B. Sprouty is a general inhibitor of receptor tyrosine kinase signaling. Development (Cambridge, England) 1999;126:4139–47. doi: 10.1242/dev.126.18.4139. [DOI] [PubMed] [Google Scholar]
- Roch F, Jimenez G, Casanova J. EGFR signalling inhibits Capicua-dependent repression during specification of Drosophila wing veins. Development. 2002;129:993–1002. doi: 10.1242/dev.129.4.993. [DOI] [PubMed] [Google Scholar]
- Rogge R, Green PJ, Urano J, Horn-Saban S, Mlodzik M, Shilo BZ, Hartenstein V, Banerjee U. The role of yan in mediating the choice between cell division and differentiation. Development Cambridge England. 1995;121:3947–3958. doi: 10.1242/dev.121.12.3947. [DOI] [PubMed] [Google Scholar]
- Rohrbaugh M, Ramos E, Nguyen D, Price M, Wen Y, Lai ZC. Notch activation of yan expression is antagonized by RTK/pointed signaling in the Drosophila eye. Current biology CB. 2002;12:576–581. doi: 10.1016/s0960-9822(02)00743-1. [DOI] [PubMed] [Google Scholar]
- Roth S, Neuman-Silberberg FS, Barcelo G, Schüpbach T. cornichon and the EGF receptor signaling process are necessary for both anterior-posterior and dorsal-ventral pattern formation in Drosophila. Cell. 1995;81:967–78. doi: 10.1016/0092-8674(95)90016-0. [DOI] [PubMed] [Google Scholar]
- Samakovlis C, Manning G, Steneberg P, Hacohen N, Cantera R, Krasnow MA. Genetic control of epithelial tube fusion during Drosophila tracheal development. Development (Cambridge, England) 1996;122:3531–6. doi: 10.1242/dev.122.11.3531. [DOI] [PubMed] [Google Scholar]
- Schober M, Rebay I, Perrimon N. Function of the ETS transcription factor Yan in border cell migration. Development Cambridge England. 2005;132:3493–3504. doi: 10.1242/dev.01911. [DOI] [PubMed] [Google Scholar]
- Scholz H, Deatrick J, Klaes A, Klämbt C, Klambt C. Genetic Dissection of pointed, a Drosophila Gene Encoding Two ETS-Related Proteins. Genetics. 1993;135:455–468. doi: 10.1093/genetics/135.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwartz A, Yogev S, Schejter ED, Shilo BZ. Sequential activation of ETS proteins provides a sustained transcriptional response to EGFR signaling. Development (Cambridge, England) 2013;140:2746–54. doi: 10.1242/dev.093138. [DOI] [PubMed] [Google Scholar]
- Sopko R, Perrimon N. Receptor tyrosine kinases in Drosophila development. Cold Spring Harbor perspectives in biology. 2013:5. doi: 10.1101/cshperspect.a009050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant MA, Roark M, Bier E. The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGF-R signaling pathway. Genes & development. 1993;7:961–73. doi: 10.1101/gad.7.6.961. [DOI] [PubMed] [Google Scholar]
- Tsachaki M, Sprecher SG. Genetic and developmental mechanisms underlying the formation of the Drosophila compound eye. Developmental dynamics : an official publication of the American Association of Anatomists. 2012;241:40–56. doi: 10.1002/dvdy.22738. [DOI] [PubMed] [Google Scholar]
- Venken KJT, He Y, Hoskins RA, Bellen HJ. P [acman]: A BAC Transgenic Platform Fragments in D. melanogaster. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Webber JL, Zhang J, Cote L, Vivekanand P, Ni X, Zhou J, Nègre N, Carthew RW, White KP, Rebay I. The relationship between long-range chromatin occupancy and polymerization of the Drosophila ETS family transcriptional repressor Yan. Genetics. 2013;193:633–49. doi: 10.1534/genetics.112.146647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Kauffmann RC, Zhang J, Kladny S, Carthew RW. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye [In Process Citation] Cell. 2000;103:87–97. doi: 10.1016/s0092-8674(00)00107-0. [DOI] [PubMed] [Google Scholar]
- Yasugi T, Sugie A, Umetsu D, Tabata T. Coordinated sequential action of EGFR and Notch signaling pathways regulates proneural wave progression in the Drosophila optic lobe. Development (Cambridge, England) 2010;137:3193–203. doi: 10.1242/dev.048058. [DOI] [PubMed] [Google Scholar]
- Zartman JJ, Cheung LS, Niepielko MG, Bonini C, Haley B, Yakoby N, Shvartsman SY. Pattern formation by a moving morphogen source. Physical biology. 2011;8:045003. doi: 10.1088/1478-3975/8/4/045003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zartman JJ, Kanodia JS, Cheung LS, Shvartsman SY. Feedback control of the EGFR signaling gradient: superposition of domain-splitting events in Drosophila oogenesis. Development. 2009;136:2903–2911. doi: 10.1242/dev.039545. [DOI] [PMC free article] [PubMed] [Google Scholar]