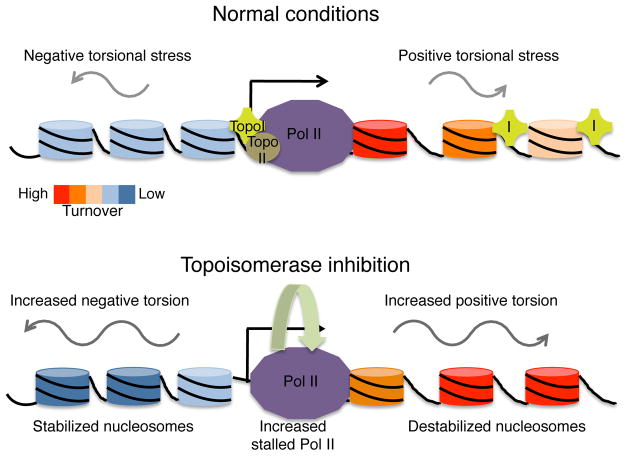
Figure 7. Model for transcription-generated torsional strain and nucleosome turnover.
Under normal conditions, RNA Polymerase II (Pol II) generates positive supercoils ahead, and negative supercoils behind, as it elongates a transcript along a gene. Topoisomerases I and II (purple cross labeled I and brown circle labeled II, respectively) mitigate the accumulation of torsional strain. Transcription results in nucleosome turnover and decondensed organization of transcribed nucleosomes. Upon topoisomerase inhibition, torsional strain, both positive and negative, accumulates, resulting in increased Pol II stalling and nucleosome destabilization, as measured by nucleosome turnover and low-salt solubility, within gene bodies. This supports a balance between destabilization of nucleosomes for Pol II passage and maintenance of chromatin structure for chromosomal integrity.