Figure 7.
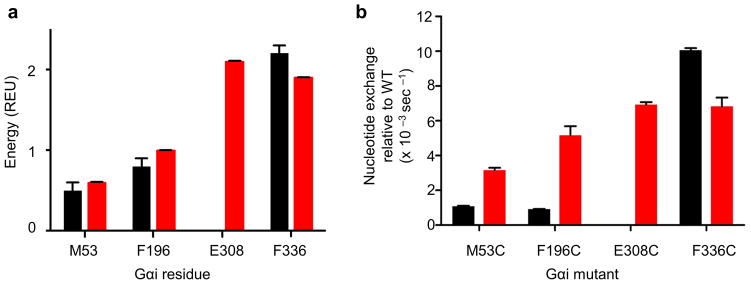
Validation of the model energetic predictions. (a) The predicted energetic contribution to a given residue’s corresponding interface is plotted for basal (black) and stimulated (red) states. Residues M53, F196, and F336 are within the α5 | Gαi interface. Residue E308 is within the R* | Gαi interface, and therefore no interface contributions are predicted in the basal state. Energy is given in Rosetta Energy Units (REU). Bars show mean, and error bars show standard deviation. (b) Basal and receptor-mediated nucleotide exchange rates. Gαi mutant exchange rates were compared, taking the absolute value of the difference of the nucleotide exchange rate relative to wild type Gαi in both basal and receptor mediated state. Data represent between 4 – 8 independent experiments, and error bars show standard error of the mean. The basal exchange of E308C was determined experimentally but was not significantly different from WT (wildtype), as was predicted by the model.
