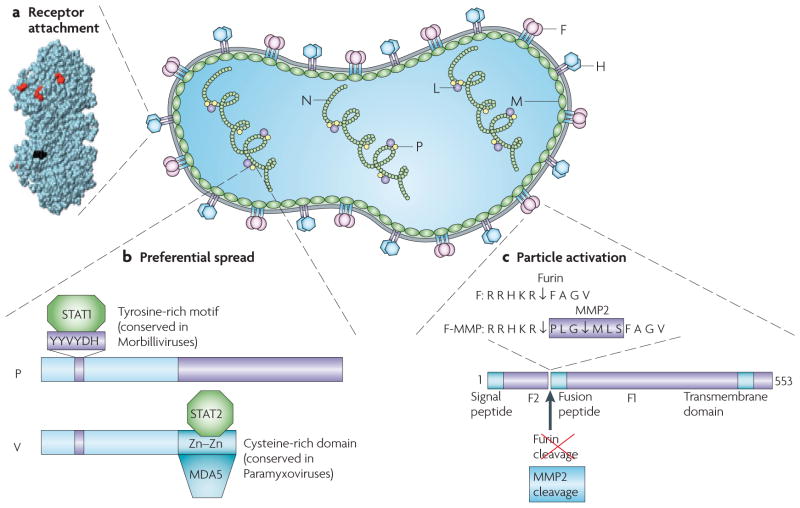
Figure 3. MV particle structure, genome organization and targeting approaches.
The MV genome has six genes that code for eight proteins. The first gene codes for the nucleocapsid protein (N) that encapsidates the genomic RNA. The last gene (L) codes for the polymerase protein that replicates and transcribes the genome together with the phosphoprotein (P), a polymerase cofactor. The matrix protein (M) organizes virus particle assembly. The two glycoproteins haemagglutinin (H) and the fusion (F) protein contact the receptor and execute membrane fusion, respectively. Two non-structural proteins that are coded by the P gene (C and V) control the innate immune response. a | Three-dimensional structure of MV H117. Residues that are necessary for signalling lymphocytic activation molecule (SLAM)-dependent or CD46-dependent fusion are in red. The site of addition of the single-chain fragment variable (scFv) is in black. b | A schematic of the P and V proteins that are encoded by the P gene. These proteins share their amino-terminal domain, but differ at the carboxyl terminus. The residues of the V and P common domain that are important for the interaction with STAT1 (signal transducer and activator of transcription 1) have been characterized. Three amino acids in a conserved hexapeptide are shown in purple. Data from the Horvath group indicate that STAT2 and MDA5 interact with different sequences in the unique cysteine-rich domain of V (A. Ramachandran, J-P. Parisien and C.M. Horvath, unpublished observations). c | Schematic of the MV F protein and amino acid sequences of its cleavage site. The standard F protein is cleaved into F1 and F2 fragments by furin, a ubiquitous protease. Furin cleavage occurs even after a hexameric peptide that codes for a matrix metalloproteinase 2 (MMP2) cleavage site is introduced (F-MMP), but the resulting F1 protein, which is extended by six residues, is inactive. Trimming of three amino-terminal residues by MMP2 cleavage confers function to F-MMP.