Abstract
Groucho related genes encode transcriptional repressor proteins critical for normal developmental processes. The Bone morphogenetic proteins belong to the transforming growth factor-β (TGF-β) superfamily and play important signaling roles in development and disease. However, the regulation of BMP signaling, especially within cells, is largely unknown. In this report, we show that expression of the Groucho related gene Grg4 robustly activates the expression of a BMP reporter gene, as well as enhancing and sustaining the upregulation of the endogenous Id1 gene induced by BMP7. BMP7 administration did not affect the endogenous level of Grg4 nor did it enhance the phosphorylation of receptor activated Smad proteins. Rather, Grg4 expression reduced the levels of the endogenous inhibitory Smad7, thus increasing the transcriptional responses mediated by BMP responsive sequences. The data point to a novel mechanisms for attenuating BMP signaling through altering the ratio of activating versus inhibitory Smad proteins.
Keywords: Groucho, BMP signaling, Smad7, Grg4, kidney fibrosis
1. Introduction
The regulation of gene expression in response to secreted signaling proteins is critical for normal cellular physiological processes, such as proliferation, differentiation, and programmed cell death. Bone morphogenetic proteins (BMPs) are secreted proteins that belong to the transforming growth factor (TGF)-β superfamily and have a broad range of biological effects [1,2]. In mammals, the binding of BMP ligands to their receptor, BMP type II receptor, leads to the recruitment and phosphorylation of BMP type I receptor (BMPRI). The activated BMPRI is a serine/threonine kinase that transduces the signal through phosphorylating receptor-activated Smad1, 5 and 8. Phosphorylated Smad1/5/8 form a heteromeric complex with a common partner, Smad4, and translocate to the nucleus to regulate target gene expression. The inhibitory Smads (I-Smads, Smad6 and 7) shared a common sequence with R-Smads and competed with them to bind to type I receptor or Smad4, thus blocking signaling transduction. In the nucleus, Smad proteins cooperate with other DNA binding proteins to activate transcription at target loci. However, the level of activation can be attenuated through the availability of co-factors, the turnover of P-Smads, and/or the levels of inhibitory Smad proteins.
One of the first repressor proteins identified in metazoans is encoded by the Drosophila Groucho (Gro) gene. Its evolutionary conserved homologues are the mammalian Groucho related genes (Grg) or Transducin-Like Enhancer of split (Tle) proteins [3]. Their structures are characterized by five domains, the Trp-Asp-repeat (WDR) domain at the N-terminus, followed by Ser-Pro-rich (SP), CcN, and Gly-Pro-rich (GP) domain, with a Gln-rich (Q) domain at the C-terminus [4]. The WDR and Q domain are highly conserved and essential for the interaction with other DNA-binding proteins and mediate gene repression [5,6]. The SP domain could be phosphorylated by MAPK, which negatively regulated Grg/Tle repression ability [7], whereas the CcN domain contains the nuclear localization signals [4]. The Grg/Tle family represses gene expression through multiple mechanisms. First, it interacts with TFIIE or other transcriptional factors to prevent the assembling of transcription machinery or activator complexes [4]. Second, Grg3 has been shown to bind nucleosomal arrays to promote condensation into higher-order chromatin that blocked the access of other transcriptional factors [8]. Third, they recruit histone deacetylase or other histone modification complex to repress target gene expression [9,10]. Through a series of knockdown and overexpression experiments, Grg/Tle proteins play important role in embryogenesis, body patterning and organogenesis [11,12,13].
BMP proteins are also critical signals in development and human disease. For example, BMP7 null mice died shortly after birth because of severe renal dysfunction [14]. BMP7 promotes the survival of metanephric mesenchymal cells, the progenitor cells of the nephron, as well as their differentiation into the diverse epithelial cells types [15,16,17]. Furthermore, BMP7 is though to be protective in several animal models of renal interstitial fibrosis and may even reverse fibrosis in some circumstances [18,19,20]. BMP signaling is regulated at multiple levels, the best studied being through interactions with other extracellular proteins that bind BMPs and/or receptors, such as Noggin [21], Gremlin [22], or KCP [23].
Both the Groucho related gene, Grg4, and BMP7 are expressed in the developing kidney and nervous system [24]. Although Grg4 can regulate diverse signaling pathways, such as Wnt, Notch and EGF signaling [3], it had not been linked to BMP or TGF-β family signaling. In this report, we find a novel interaction between Grg4 and BMP signaling. Using a reporter system that responds to BMPs, we find that Grg4 activates genes that are driven by BMP responsive elements (BREs). This activation is observed even when a known Grg4 mediated repressor sequence is adjacent to the BREs. Since Grg4 does not bind to DNA directly, we investigated the mechanisms of Grg4 mediated activation of the BMP pathway. We find that activation of the BREs and of the endogenous BMP target gene, Id1, is through Grg4 dependent repression of the inhibitory Smad7 protein. These data demonstrate how a repressor can activate a cell signaling pathway by altering the balance of receptor activated Smad and inhibitory Smad proteins.
2. Materials and Methods
2.1 Reporter Molecular Construction
The forward and reverse strands of the BRE sequence were synthesized with BamHI site at the 5’-ends and cloned into the BamHI site of pRS4-EGFP reporter vector. To delete the Pax2 binding sites from Pax2 and BMP double reporter vector, the vector was digested by HindIII and EcoRV overnight, blunt ended by the Klenow Fragment (NEB), and re-ligated. BRE fragment sense: 5’-GATCCGCGGCGCCAGCCTGACAGCCCGTCCTGGCGTCTAACGGTCTGAGCTAGCG-3’; reverse: 5’-GATCCGCTAGCTCAGACCGTTAGACGCCAGGACGGGCTGTCAGGCTGGCGCCGCG-3’.
2.2 Cell Culture
HEK293 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM, Gibco) with 10% Fetal Bovine Serum (FBS), and Penicillin Streptomycin (PS, Gibco). Immortalized renal epithelial cells (TKPTS) were a kind gift from Dr. Bello-Reuss. Cells were cultured in Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F-12, Gibco) with 2% FBS, 1× Insulin-Transferring-Ethanolamine-Selenium (ITES, Lonza) and PS. UltraMDCK serum free medium (Lonza) was used when serum starvation was necessary.
To test the effect of Pax2, Grg4 or BMP7 on reporter vectors, 293 cells were culture on 6 well plates with low serum medium (LSM, DMEM+0.5% FBS+1× Insulin-Transferring-Selenium (ITS, Gibco)) and transfected with 0.5µg reporter vectors and 0.5–1 µg Pax2, Grg4 expressing vector, or SHS (sonicated herring sperm) DNA as control, using Fugene6 (Roche). Cells were harvest 48 hours after transfection for analysis. To test the effect of Smad7 on BMP reporters, cells were transfected with 0.5µg pRS4-BRE4+EGFP reporter vector, 0.5µg Grg4 expressing vector, and 0.5µg Smad7 expressing vector or SHS DNA control. 100 ng/mL BMP7 (R&D systems) was added 24 hour after transfection for another 24 hours. For 1 hour pulse experiment, transfected cells were treated with 100 ng/mL BMP7 for 1h, and then washed with PBS once and cultured in new LSM for another 23 hours.
To collect the conditioned medium, 293 cells were cultured on 100 mm dish and transfected with 5 µg of GFP or Grg4 expressing vector, using Fugene6. 48 hours after transfection, culture medium from each plate was collected and centrifuged at 4000 rpm for 30 min at 4°C. The supernatant was aliquot and preserved in −80°C.
2.3 Western blot analysis
Cells were directly lysed in 2× SDS buffer (4% sodium dodecyl sulfate, 20% glycerol, 0.2M dithiothreitol, 125 mM Tris, pH 6.8) and boiled at 94°C. Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to PVDF membranes and immunoblotted with antibodies as indicated. Rabbit anti-phosphorylated Smad1/5/8 is from Cell Signaling. Mouse anti-flag and mouse anti β-tubulin are from Sigma-Aldrich. Mouse anti-EGFP, mouse anti-Smad1 and rabbit anti-Tle4 are from Santa Cruz Biotech. Rabbit anti-Pax2 is described [25]. HRP-linked secondary antibodies and ECL reagent are from GE healthcare.
2.4 RNA reverse-transcription and real-time PCR
Total RNA was extracted from 293 cells with different treatment using TRIZOL RNA isolation system (Invitrogen). 2–3 µg total RNA was reverse-transcribed into complementary DNA with SuperScript First-Strand Kit (InVitrogen). The cDNA products were diluted 5 times and amplified with the iTaq Sybrand cultured for 24 hours green master mix (Bio-Rad) in a Prism 7500 (Applied Biosystems). Primers pairs for PCR are as follows: Id1 5’-CTGCCTGCCCTGCTGGAC-3’, 5’-TCTCGCCGTTGAGGGTGC-3’; Tle4 5’-TACCCCTACTCCACGAACT-3’, 5’-TCTCCGTTCATTCCAGCA-3’; Smad4 5’-CACTACGAACGAGTTGTATCAC-3’, 5’-CCTTCAGTGGACAACGATG-3’; Smad7 5’-ATCACCTTAGCCGACTCTG-3, 5’-CAGTAGAGCCTCCCCACTC-3’; L32 5’- CAGGGTTCGTAGAAGATTCAAGGG-3’, 5’-CTGGAGGAAACATTGTGAGCGATC-3’.
2.5 Luciferase assays
293 cells were seeded on 12 well plates and cultured in LSM. BRE-luc reporter vector was transfected (1 µg/well) together with SHS or Tle4 expressing vector (1 µg/well) into the cells in triplicate. Medium containing 100ng/mL BMP7, or GFP or Tle4 conditional medium was added 24 hours after transfection and kept for another 24 hours. Cells were lysed with dual luciferase assay kit (Promega) and results were read.
2.6 shRNA mediated Gene knocking-down
Packed Smad4 37196 or 37199 shRNA lentivirus was used to knockdown in PRECs. Cells were seeded on 6 well plate for 24 hours. Lentivirus was added with 8 µg/mL polybrene and kept overnight. Puromycin was added and kept for consistent selection. For the BMP reporter test in Smad4 knockdown cells, cells were seeded on 12 well plate and cultured for 24 hours. 1.5 µg of DNA, containing 0.5 µg of pRS4-BRE4+-EGFP reporter and 1µg of Tle4 expressing vector or SHS DNA control was transfected using Fugene6. 48 hours later, cells were lysed in 2×SDS loading buffer and analyzed by western blotting.
3. Results
Molecular Construction of BMP reporter vectors
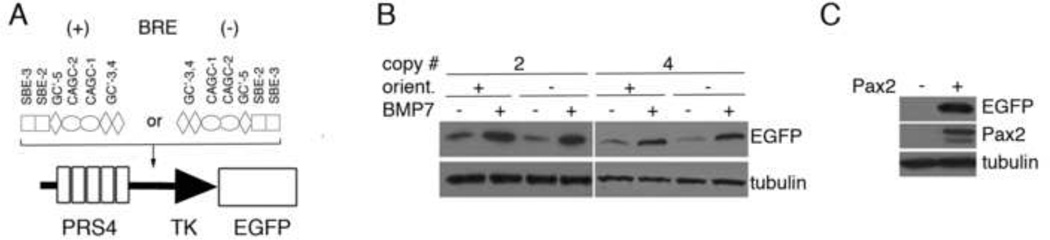
The Groucho proteins are recruited to chromatin through interactions with DNA binding proteins. Previously, we have shown that Grg4 recruitment by the Pax2 DNA binding protein inhibits gene expression by displacing the co-activator PTIP and subsequently recruiting the Polycomb repressor 2 complexes (PRC2) [10]. However, promoter and enhancer sequences rarely bind just one factor, rather they constitute regions where multiple activators or repressor bind. We used the Pax2 reporter system to ask whether Grg4 mediated repression is dominant, within the context of another activating sequence. In other words, can Grg4 mediated recruitment of PRC2 suppress another activation sequence when placed adjacent to a Pax2 binding site in cis? To test this hypothesis, we inserted a strong BMP response element (BRE) between the Pax2 response sequence (PRS4) and a minimal TK promoter that drives enhanced green fluorescent protein (Fig. 1A). The BRE was derived from sequences found at the Id1 gene promoter and is strongly activated by BMP7 in most cells [26]. Multiple copies of the BRE were inserted in either direction and tested for their ability to respond to BMP7 by transient transfection. Both 2 or 4 copies of the BRE were able to drive EGFP expression upon addition of BMP7, regardless of the orientation (Fig. 1B). The presence of the BRE element also did not affect the ability of Pax2 to also stimulate EGFP expression when both elements were present (Fig. 1C).
Figure 1. Construction of BMP and Pax2 Double Reporter Vectors.
A) The Bmp Response Element (BRE) was inserted between a Pax2 Response sequence (PRS4) and a minimal HSV Thymidine Kinase (TK) promoter and the Enhanced Green Fluorescent Protein coding region. The orientations were designated as + or −. Specific sequence elements within the BRE were as described [26]. B) Transient transfection of BMP/Pax2 reporters containing 2 or 4 copies of the BRE in either orientation followed by addition of Bmp7 (100 ng/ml) and western blotting for EGFP 48h post transfection. Note Bmp7 dependent activation of EGFP expression. C) Co-transfection of Bmp7/Pax2 reporter with or without Pax2 expression plasmids shows that EGFP activation is still Pax2 dependent in the absence of Bmp7.
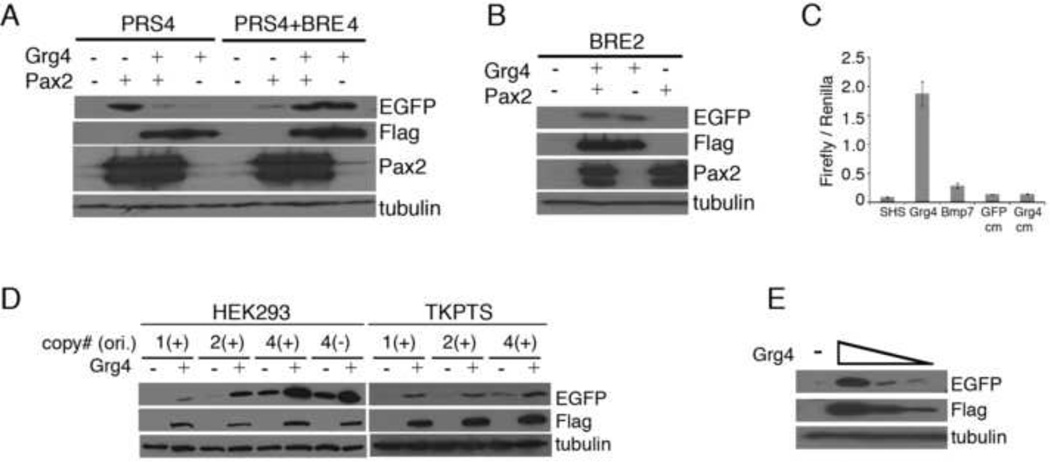
We then tested whether Grg4/Pax2 could suppress BMP mediated activation when both the BRE and the PRS4 sequences were present. As reported previously, Grg4 expression inhibited activation by Pax2 when the PRS4 element was used alone (Fig. 2A). However, co-expression of Grg4 and Pax2 did not suppress the EGFP reporter gene when the BRE elements were positioned between the PRS4 sequences and the TK minimal promoter (Fig. 2A). Surprisingly, reporter gene expression was higher when Grg4 was present than with Pax2 alone. When Grg4 was expressed by itself, activation of EGFP was just as strong as with Pax2/Grg4. Not only was the Pax2/Grg4 mediated repression lost, but Grg4 activated the PRS4-BRE element although Grg4 had no effect on the PRS4 sequences alone. These data suggested that Grg4 could act upon the BRE and that this function was activating, not suppressing, the downstream reporter.
Figure 2. Activation of the BRE Reporter Plasmid by Grg4.
A) Pax2 reporter plasmid (PRS4) or pPax2/Bmp7 double reporter plasmids (PRS4+BRE) were co-transfected with Pax2 or with Pax2 and flag-Grg4. EGFP levels were assayed by western blotting. Note that Grg4 alone activates the double reporter but suppresses Pax2 activation of the single reporter (PRS4). B) The BRE element is sufficient to mediate Grg4 dependent activation. Two copies of the BRE element were inserted upstream of the TK-EGFP plasmid and co-transfected with either Pax2 or flag-Grg4 and expression of EGFP analyzed by western blotting. Note that Grg4 activates EGFP but in the absence of the PRS4 element Pax2 does not activate. C) Luciferase assays using the BRE-Luc reporter and co-transfection with flag-Grg4 or the addition of recombinant Bmp7. Conditioned media (cm) from GFP or Grg4 transfected cells was also tested for activation of BRE-Luc. Note that Bmp7 activates BRE-Luc approximately 3–4 fold but Grg4 expression activates more than 15 fold. D) Copy number and orientation of the BRE in transfected HEK293 or renal epithelial TKPTS cells. Copy number affects base level EGFP reporter expression but does not significantly impact Grg4 mediated activation. E) Increasing amounts of transfected Grg4 show increased levels of EGFP activation using the BRE-EGFP reporter.
The BRE elements were then tested directly for activation by Grg4 in the absence of the PRS4 sequences using our EGFP reporter (Fig. 2B) or the BRE-luc reporter (Fig. 2C) described previously. In both HEK293 cells and renal epithelial cells, expression of Grg4 activated reporter gene expression when driven by the BRE. The Grg4 mediated activation and the level of EGFP expression increased when multiple copies of the BREs were utilized (Fig. 2D). Furthermore, EGFP activation was proportional to the amounts of Grg4 transfected (Fig. 2E). These data suggested that Grg4 may be binding to or interacting with the BRE elements, perhaps through another DNA binding protein, such as the BMP effector Smads 1/5/8. Although Grg4 could bind to the PRS4 sequence when Pax2 was present, we could find no evidence of Grg4 binding to the BRE sequence, either directly or indirectly, by chromatin immunoprecipitation (data not shown).
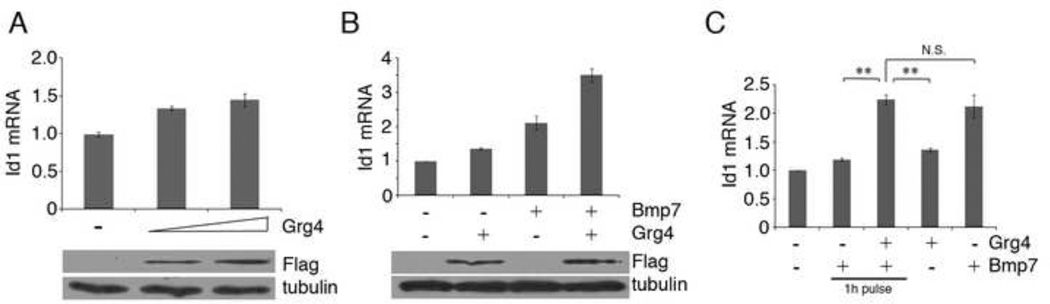
To examine whether the Grg4 dependent activation of EGFP was unique to our constructs or represented a real biological phenomenon, we examined the expression of the endogenous Id1 gene is response to Grg4, BMPs, or both. At the highest doses, expression of Grg4 stimulated Id1 mRNA by 1.5 fold over controls (Fig. 3A), whereas BMP7 addition resulted in a 2 fold increase of Id1 mRNA (Fig. 3B). However, Grg4 and BMP together increased Id1 mRNA levels nearly 4 fold (Fig. 3B). The expression of Grg4 also enhanced Id1 activation when only a short pulse of BMP7 was given, followed by a chase with fresh media. These data indicate that expression of Grg4 sensitized cells to BMP7 and enhanced the expression of target genes.
Figure 3. Activation of Endogenous Id1 mRNA Expression.
A) RT-PCR of Id1 mRNA from HEK293 cells transfected with increasing amounts of Grg4. B) RT-PCR of Id1 mRNA after addition of Bmp7 and transient transfection of Grg4 as indicated. Note that Bmp7 and Grg4 show maximal levels of Id1 activation. C) Transient transfection of Grg4 sensitizes cells to Bmp7 treatment. Cells were transfected with Grg4 and then exposed to a 1h pulse of Bmp7 or continuous Bmp7 exposure for 24h. Note that Id1 mRNA levels were significantly increased with a 1h Bmp7 pulse only after Grg4 expression.
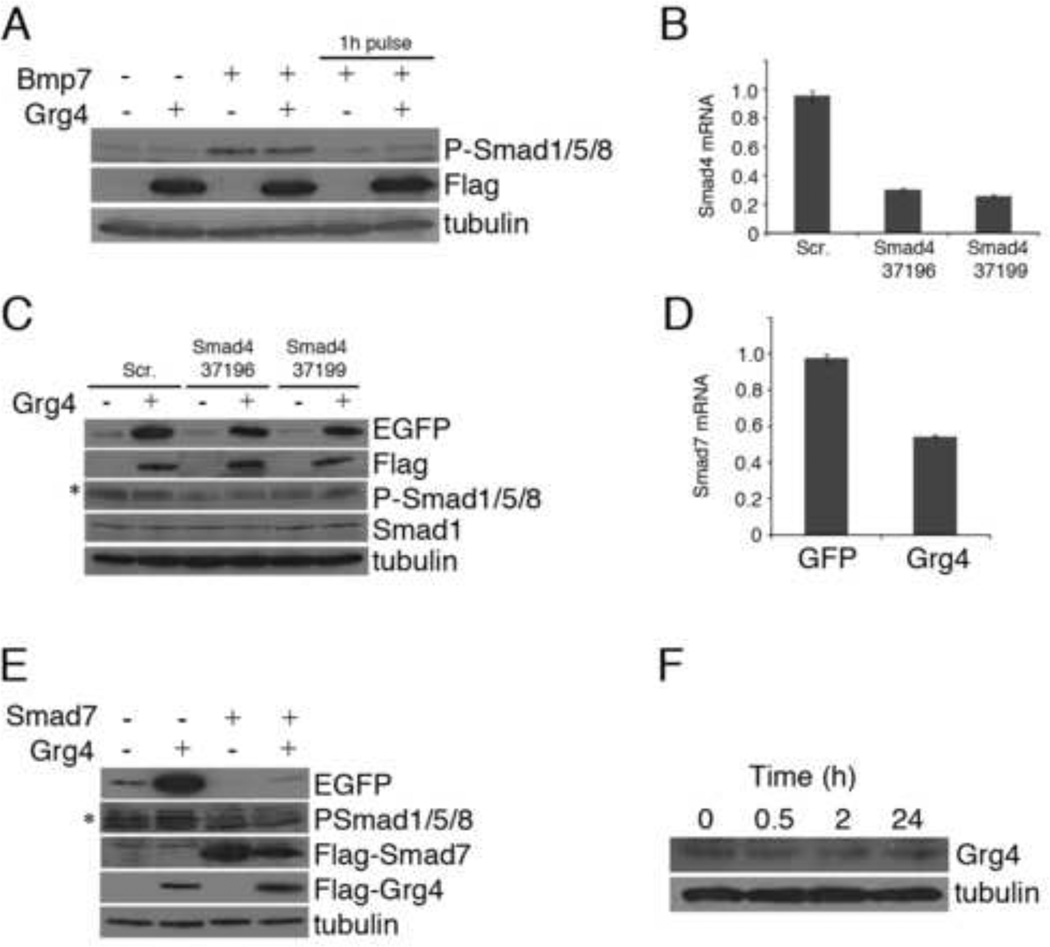
If Grg4 does not directly bind to the BREs, what might be the mechanisms of BRE activation? One possibility is that Grg4 increases levels of P-Smad1/5/8, perhaps through the activation of endogenous BMP proteins or suppression of an inhibitor. However, we found no evidence of increased P-Smad1/5/8 levels (Fig. 4A). Expression of Grg4 alone, or in combination with BMP7 did not alter the levels of P-Smad1/5/8. We also examined whether altering the levels of Smad4 affected the Grg4 mediated activation (Fig. 4B, C). Despite reducing Smad4 mRNA levels to 30% of normal, we did not observe a reduction of Grg4 mediated reporter gene activation. HEK293 cells also express the inhibitory Smad7 protein, so we next examined whether Grg4 could affect levels of Smad7. Strikingly, expression of Grg4 inhibited endogenous levels of Smad7 mRNA by 50% (Fig. 4D). Furthermore, co-transfection with Smad7 expression vectors completely abrogated the Grg4 mediated activation of the EGFP reporter gene. Lastly, we examined whether BMP7 could activate endogenous Grg4 to enhance signaling by inhibiting Smad7. However, no significant effects were observed on Grg4 protein levels by the addition of BMP7. These data indicate that Grg4 has the potential to enhance BMP signaling and increase gene expression of BMP responsive genes by reducing the levels of the inhibitory Smad7 protein.
Figure 4. Grg4 mediated activation through inhibition of Smad7.
A) Grg4 does not affect BMP mediated phosphorylation of Smad1/5/8. Cells were transfected with or without Grg4 and cultured with Bmp7 for 24h or a 1h pulse followed by 24h chase. Western blots indicate the levels of P-Smad1/5/8 and flag-Grg4. B) SiRNAs against Smad4 reduce levels of SMad4 mRNA. C) Smad4 knockdown does not affect EGFP reporter gene expression in response to Grg4. Western blots show levels of EGFP, flag-Grg4, P-Smad1/5/8 and tubulin loading controls. D) Quantitative RT-PCR of Smad7 mRNA in control (GFP) or Grg4 transfected cells. E) Co-transfection of Smad7 inhibits the Grg4 mediated EGFP reporter gene expression. F) Endogenous levels of Grg4 are not affected in BMP7 treated HEK293 cells.
4. Discussion
The Grg/Tle family proteins are common co-repressors that can impact a variety of signaling pathways in development. For example, Tle proteins competed with β-catenin to interact with Tcf/Lef, thus inhibiting canonical Wnt signaling [27]. In Drosophila, the downstream effector of the decapentaplegic signaling, brinker, recruits Groucho and CtBP to suppress specific target genes [28]. Previously, we have shown that expression of Grg4 is regulated during development of the kidney and nervous system and partially overlaps with cells and tissues that respond to BMP signals [24]. In concert with the DNA binding protein Pax2, Grg4 is able to recruit histone methyltransferases and PRC2 to chromatin and inhibit gene expression [10].
In this report, we used a BMP response element to test whether Grg4 mediated repression at an adjacent sequence could affect the ability of BMPs to activate a target gene driven by BREs. To our surprise, we found that Grg4 has no repressive effects when the BREs are present. On the contrary, Grg4 is able to activate gene expression when the reporter gene is driven by the BRE sequences only. Since we could not localize Grg4 to the BRE sequences, we examined alternative mechanisms for Grg4 mediated activation. Given that Grg4 suppressed endogenous levels of the inhibitory Smad7 and that over-expression of Smad7 inhibited Grg4 mediated activation of BMP targets, we conclude that Grg4 dependent activation of the BMP pathway is most likely mediated by a reduction of inhibitory Smads.
Groucho proteins are potent transcriptional repressors that have been linked to the recruitment of Polycomb repressor complexes and chromatin condensation [8, 10]. How this repressor function of Gro/Tle proteins could modify TGF-β superfamily signaling pathways was largely unexplored. To date, it was only reported that Dpp, the TGF-β homolog in Drosophila, induced the expression of Brinker, which recruited Groucho and CtBP to repress other Dpp target genes, thus confining the actions of Dpp signaling to a particular region or active zone [28,29]. However, this required direct recruitment of Groucho to Dpp target genes. In our experimental system, we show that Grg4 can modify BMP signaling not by any direct interactions with the BRE DNA sequences, but by repression of inhibitory proteins that attenuate the BMP signaling response. This represents a potentially novel mechanism for fine-tuning the BMP signal in development and in adult cells.
Smad7 can inhibit BMP signaling responses by interfering with the binding of R-Smads to Smad4 or to the type I receptor [30]. Yet in our Smad4 knockdown experiment, Grg4 still activated the BMP reporter. However, not all TGF-β family signal responses require Smad4, as previous studies identified some Smad4 independent target genes [31,32]. Our data suggested that by reducing Smad7 protein levels, sufficient endogenous BMPs and basal levels of P-Smad1/5/8 are present to increase activation of genes driven by the BREs or related sequences.
In summary, the regulation of BMP7 signaling is critical for renal development and renal disease progression. The present data showed that Grg4 can play an important role in regulating BMP7 mediated activation of reporter and endogenous genes. By altering the levels of inhibitory Smads, Grg4 can enhance BMP signaling and potentially provide novel mechanisms for enhancing the therapeutic effects of BMPs in fibrotic disease.
Highlights.
The repressor protein Grg4 activates transcription of BMP target genes.
Activation is mediated by the BMP response elements.
Levels of P-Smad or co-activator Smads do not correlate with Grg4 activation.
Grg4 reduces the levels of the inhibitory Smad7 to promote BMP mediated activation.
Acknowledgments
This work was supported by National Institutes of Health Grants DK054740 and DK062914 to G.R.D.
Abbreviations
- Grg4
Groucho related gene 4
- Tle
Transducin-like enhancer of split
- BMP
bone morphogenetic protein
- TGF
transforming growth factor
- BRE
Bmp Response element
- PRC2
Polycomb repressor complex 2
- EGFP
enhanced green fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
The authors declare that they have no competing financial interests.
References
- 1.Patel SR, Dressler GR. BMP7 signaling in renal development and disease. Trends Mol Med. 2005 doi: 10.1016/j.molmed.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 2.Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. Embo J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cinnamon E, Paroush Z. Context-dependent regulation of Groucho/TLE-mediated repression. Curr Opin Genet Dev. 2008;18:435–440. doi: 10.1016/j.gde.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Buscarlet M, Stifani S. The 'Marx' of Groucho on development and disease. Trends in Cell Biology. 2007;17:353–361. doi: 10.1016/j.tcb.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Jennings BH, Pickles LM, Wainwright SM, Roe SM, Pearl LH, Ish-Horowicz D. Molecular recognition of transcriptional repressor motifs by the WD domain of the Groucho/TLE corepressor. Mol Cell. 2006;22:645–655. doi: 10.1016/j.molcel.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 6.Fisher AL, Caudy M. Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 7.Hasson P, Egoz N, Winkler C, Volohonsky G, Jia S, Dinur T, Volk T, Courey AJ, Paroush Z. EGFR signaling attenuates Groucho-dependent repression to antagonize Notch transcriptional output. Nature Genetics. 2005;37:101–105. doi: 10.1038/ng1486. [DOI] [PubMed] [Google Scholar]
- 8.Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yochum GS, Ayer DE. Pf1, a novel PHD zinc finger protein that links the TLE corepressor to the mSin3A-histone deacetylase complex. Molecular and Cellular Biology. 2001;21:4110–4118. doi: 10.1128/MCB.21.13.4110-4118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel SR, Bhumbra SS, Paknikar RS, Dressler GR. Epigenetic mechanisms of Groucho/Grg/TLE mediated transcriptional repression. Molecular Cell. 2012;45:185–195. doi: 10.1016/j.molcel.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Wang YG, Reginato AM, Glotzer DJ, Fukai N, Plotkina S, Karsenty G, Olsen BR. Groucho homologue Grg5 interacts with the transcription factor Runx2-Cbfa1 and modulates its activity during postnatal growth in mice. Developmental Biology. 2004;270:364–381. doi: 10.1016/j.ydbio.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Zamparini AL, Watts T, Gardner CE, Tomlinson SR, Johnston GI, Brickman JM. Hex acts with beta-catenin to regulate anteroposterior patterning via a Groucho-related co-repressor and Nodal. Development. 2006;133:3709–3722. doi: 10.1242/dev.02516. [DOI] [PubMed] [Google Scholar]
- 13.Dasen JS, Martinez Barbera JP, Herman TS, Connell SO, Olson L, Ju B, Tollkuhn J, Baek SH, Rose DW, Rosenfeld MG. Temporal regulation of a paired-like homeodomain repressor/TLE corepressor complex and a related activator is required for pituitary organogenesis. Genes and Development. 2001;15:3193–3207. doi: 10.1101/gad.932601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo G, Hofmann C, Bronckers ALJJ, Sohocki M, Bradley A, Karsenty G. BMP-7 is an inducer of nephrogenesis, and is also required for eye development and skeletal patterning. Genes and Development. 1995;9:2808–2820. doi: 10.1101/gad.9.22.2808. [DOI] [PubMed] [Google Scholar]
- 15.Dudley AT, Godin RE, Robertson EJ. Interaction between FGF and BMP signaling pathways regulates development of metanephric mesenchyme. Genes Dev. 1999;13:1601–1613. doi: 10.1101/gad.13.12.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dressler GR. The cellular basis of kidney development. Annu Rev Cell Dev Biol. 2006;22:509–529. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- 17.Vukicevic S, Kopp JB, Luyten FP, Sampath TK. Induction of nephrogenic mesenchyme by osteogenic protein 1 (bone morphogenetic protein 7) Proc Natl Acad Sci U S A. 1996;93:9021–9026. doi: 10.1073/pnas.93.17.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugimoto H, LeBleu VS, Bosukonda D, Keck P, Taduri G, Bechtel W, Okada H, Carlson W, Jr, Bey P, Rusckowski M, Tampe B, Tampe D, Kanasaki K, Zeisberg M, Kalluri R. Activin-like kinase 3 is important for kidney regeneration and reversal of fibrosis. Nat Med. 2012;18:396–404. doi: 10.1038/nm.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeisberg M, Bottiglio C, Kumar N, Maeshima Y, Strutz F, Muller GA, Kalluri R. Bone morphogenic protein-7 inhibits progression of chronic renal fibrosis associated with two genetic mouse models. Am J Physiol Renal Physiol. 2003;285:F1060–F1067. doi: 10.1152/ajprenal.00191.2002. [DOI] [PubMed] [Google Scholar]
- 20.Zeisberg M, Hanai J, Sugimoto H, Mammoto T, Charytan D, Strutz F, Kalluri R. BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med. 2003;9:964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
- 21.Zimmerman LB, De Jesus-Escobar JM, Harland RM. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 22.Michos O, Goncalves A, Lopez-Rios J, Tiecke E, Naillat F, Beier K, Galli A, Vainio S, Zeller R. Reduction of BMP4 activity by gremlin 1 enables ureteric bud outgrowth and GDNF/WNT11 feedback signalling during kidney branching morphogenesis. Development. 2007;134:2397–2405. doi: 10.1242/dev.02861. [DOI] [PubMed] [Google Scholar]
- 23.Lin J, Patel SR, Cheng X, Cho EA, Levitan I, Ullenbruch M, Phan SH, Park JM, Dressler GR. Kielin/chordin-like protein, a novel enhancer of BMP signaling, attenuates renal fibrotic disease. Nat Med. 2005;11:387–393. doi: 10.1038/nm1217. [DOI] [PubMed] [Google Scholar]
- 24.Cai Y, Brophy PD, Levitan I, Stifani S, Dressler GR. Groucho suppresses Pax2 transactivation by inhibition of JNK-mediated phosphorylation. The EMBO journal. 2003;22:5522–5529. doi: 10.1093/emboj/cdg536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dressler GR, Douglass EC. Pax-2 is a DNA-binding protein expressed in embryonic kidney and Wilms tumor. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:1179–1183. doi: 10.1073/pnas.89.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korchynskyi O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 27.Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nature structural & molecular biology. 2005;12:364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 28.Hasson P, Muller B, Basler K, Paroush Z. Brinker requires two corepressors for maximal and versatile repression in Dpp signalling. The EMBO journal. 2001;20:5725–5736. doi: 10.1093/emboj/20.20.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Levine M, Ashe HL. Brinker is a sequence-specific transcriptional repressor in the Drosophila embryo. Genes Dev. 2001;15:261–266. doi: 10.1101/gad.861201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone MA, Jr, Wrana JL, Falb D. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 31.Descargues P, Sil AK, Sano Y, Korchynskyi O, Han G, Owens P, Wang XJ, Karin M. IKKalpha is a critical coregulator of a Smad4-independent TGFbeta-Smad2/3 signaling pathway that controls keratinocyte differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2487–2492. doi: 10.1073/pnas.0712044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]