Abstract
The Human Genome Project, coupled with rapidly evolving high throughput technologies, has opened the possibility of identifying heretofore unknown biological processes underlying human disease. Due to the opaque nature of HIV-associated neurocognitive disorder (HAND) neuropathogenesis, the utility of such methods has gained notice among NeuroAIDS researchers. Further, the merging of genetics with other research areas has also allowed for application of relatively nascent fields, such as neuroimaging genomics and pharmacogenetics, to the context of HAND. In this review, we detail the development of genetic, transcriptomic, and epigenetic studies of HAND, beginning with early candidate gene association studies and culminating in current “omics” approaches that incorporate methods from systems biology to interpret data from multiple levels of biological functioning. Challenges with this line of investigation are discussed; including the difficulty of defining a valid phenotype for HAND. We propose that leveraging known associations between biology and pathology across multiple levels will lead to a more reliable and valid phenotype. We also discuss the difficulties of interpreting the massive and multi-tiered mountains of data produced by current high-throughput omics assays, and explore the utility of systems biology approaches in this regard.
Keywords: NeuroAIDS, HIV-associated neurocognitive disorder, weighted gene co-expression network analysis, WGCNA, HAND, HIV
1. Introduction
The first cases of Acquired Immunodeficiency Syndrome (AIDS) that came to the attention of western doctors over three decades ago frequently presented as rapidly advancing dementia. The dementia syndrome associated the Human Immunodeficiency Virus-1 (HIV), was first systematically described and termed AIDS Dementia Complex in 1986 1. Since that time, various other terms have been used to describe this syndrome, including HIV dementia, AIDS-related dementia, and subacute encephalitis. A systematic terminology was eventually adopted by the World Health Organization in 1990 and by the American Academy of Neurology (AAN) in 1991, coining HIV-1-associated cognitive/motor complex (HACMC) to describe the cognitive and motor syndromes associated with AIDS, and differentiating the more mild HIV-1-associated minor cognitive/motor disorder (MCMD) from the more severe HIV-1-associated dementia (HAD). Since 1991 there has been increasing evidence for a more mild form of neurocognitive impairment 2, leading to updated research criteria that captures the full spectrum of neurocognitive deficits due to HIV 3. The term for this broad classification is HIV-associated neurocognitive disorders, or HAND.
Despite the recent refinement of diagnostic criteria, controversy remains. Some argue that the most mild form of HAND, asymptomatic neurocognitive impairment (ANI), is over diagnosed4. This is partly due to the psychometric methodology used for arriving at a diagnosis. Specifically, the diagnosis of ANI requires that performance on two or more neurocognitive domains be greater than one standard deviation below the average, with no functional (e.g., cooking, managing finances) deficits. However, even supposedly healthy individuals will demonstrate variability in neurocognitive performance5, and the various methods for deriving composite scores result in marked differences in diagnostic classification6. As such, a significant proportion of HIV+ individuals diagnosed with ANI may not have HAND. Another reason for controversy is the low reliability of the HAND diagnosis. A study conducted with the National NeuroAIDS Tissue Consortium 7 showed that there is little agreement among experienced neuropsychologists regarding the cause of neurocognitive impairment among HIV+ individuals8. While the National NeuroAIDS Tissue Consortium cohort consists of individuals with advanced disease who often have comorbidities such as substance abuse, opportunistic infections of the CNS and other conditions that could confound diagnosis, the cohort also largely reflects the current nationwide demographics of the HIV pandemic. Finally, and perhaps owing largely to the two points above, there is no reliable biological marker for HAND in the current era. Whereas HIV-encephalitis (HIVE), which includes a variety of specific neuropathological markers, was frequently associated with HAD prior to the widespread advent of combined antiretroviral therapy (cART), today no such neuropathological indicators exist, although some hold promise9,10.
In recent years, studies of genomic and transcriptomic factors underlying symptoms and disease have led to remarkable insights about HAND neuropathogenesis, and have fueled optimism for identifying treatment targets. However, while the application of global and high throughput ‘omics methods coupled with bioinformatics and systems biology is promising, it faces a serious hurdle; the lack of a reliable phenotype for HAND. How can we, without a reliable neurocognitive, neuropathological, or neurophysiological phenotype for HAND, apply these methods in an effective manner? In this review, we present the current state of research utilizing human genetic, gene expression, and epigenetic data to understand HAND neuropathogenesis. We employ the term HAND to include all HIV-related neurocognitive deficits and their putative neuropathological causes. The benefits and limitations of these methods as applied to HAND are discussed. Finally, we propose potential solutions to overcome the primary obstacle in this research area; namely, a shift from behavioral to biological phenotypes, and the application of systems biology to ‘omics data as a path towards understanding the complexities of this disease process.
2. Genetic studies of HAND
2.1. Candidate Gene Studies
There has long been interest in the role of host-genetics in relation to psychiatric and neurologic illness. With regards to HAND, there are no heritable neurocognitive deficits or neuropsychiatric symptoms that would provide a foothold from which to explore disease etiology. However, variants of genes involved in various biological processes can significantly impact risk of neurocognitive impairment, disease course, and response to antiretroviral medications (ARVs). Host genetic factors have received increasing attention in the realm of HAND both as a means to identify risk factors and to help delineate the neuropathogenesis. Table 1 lists the existing studies that focused on neurocognitive dysfunction or neuropathology. Below we provide a more detailed description of those alleles most frequently examined and, in some instances, validated across studies.
Table 1.
Summary of candidate gene and genome-wide association studies of HIV-related neurocognitive disorders.
| Study 1st author (year) | Sample Description | Phenotype | Findings* |
|---|---|---|---|
| Dunlop et al., 1997 80 | 132 adult AIDS patients from Norway; majority were male Postmortem study of the basal ganglia, frontoparietal cortex, cerebral white matter, cerebellum, brain stem, thoracic spinal cord, and hippocampus. Study compared the relationship of ApoE genotype with dementia and HIVE. |
HIV dementia was rated on a graded scale by a physician (no dementia; possible dementia; clinical dementia); however, the criteria were not specified. Determination of HIVE appeared to be based on presence of multinucleated cells, microglial noduli and/or diffuse damage of white matter. Authors referred to a 1995 paper231 |
No association between ApoE genotype (rs429358 and rs7412) with either HIV dementia or HIVE |
| Corder et al. 1998 68 | 44 HIV+ adults; majority were male and Caucasian; Participants evaluated for neurological and other symptoms twice yearly for up to 10 visits |
Dementia (described as predominantly mild). Criteria were not specified. Reported use of a battery of neuropsychological tests |
ApoE ε4 allele carriers were twice as likely to be diagnosed with dementia during the study. The combination of the 4 allele and low CD4+ increased risk for dementia over time. ε4 allele carriers were also more likely to have peripheral neuropathy. |
| Sato-Matsumura et al. 1998 222 |
44 AIDS patients with autopsy-verified HIVE or HIV- leukoencephalopathy, 30 AIDS patients without these neuropathologies. |
HIVE and/or HIV- leukoencephalopahy |
TNF-2 genotype (rs# unspecified) did not differ between the two groups. |
| Boven et al. 1999 29 | 9 clinically demented AIDS patients; 8 non- demented AIDS patients; 6 HIV− control patients; age and ethnicity not provided; none of the AIDS patients were being treated with antiretroviral therapy. Post-mortem tissue specimens from the frontal cortex analyzed. |
AIDS Dementia Complex/HIV- Associated Dementia determined by a physician. Dementia cases had a score of 2 or higher on the Memorial Sloan-Kettering scale. Diagnoses were confirmed by postmortem neuropathological examination (methods not specified). |
The Delta 32 deletion variant in the CCR5 gene (rs333) was not found at the expected frequency among ADC patients. In vitro study showed heterozygosity of the Delta 32 deletion was associated with lower viral replication. |
| Van Rij et al. 1999 30 | 49 patients with AIDS Dementia Complex (ADC); 186 AIDS patients who died of AIDS with no ADC; age and ethnicity not provided; none received triple antiretroviral therapy |
AIDS Dementia Complex- criteria were not specified |
Lower frequency of the Delta 32 deletion variant in the CCR5 gene (rs333) among ADC patients. CCR2 64I (rs1799864) genotype did not differ between ADC and none-ADC patients. |
| Quasney et al. 2001 55 | 16 HIV+ adults with dementia, 45 HIV+ adults without dementia, and 231 healthy adult controls; 45-56% were Caucasian |
HIV dementia per AAN criteria. The Memorial Sloan-Kettering criteria was used to classify severity 232 |
TNF-α-308 (rs1800629) A allele was significantly over- represented among those with HIV dementia |
| Gonzalez et al. 2002 39 | 1) 1151 HIV+ ethnically diverse adults (55% European-American, 36% African-American, 6% Hispanic-American, and 3% other). The majority was male (94%). Sample was followed prospectively with a median follow-up time of 5.9 years 2) 592 Argentinean children perinatally exposed to HIV (322 HIV+, 270 HIV-); drawn from a larger prospective study with a median follow-up time of 4.08 years |
HIV Associated Dementia. Criteria were derived from the Center for Disease Control233, and included neuropsychological testing and neuroimaging. |
In Caucasian adults, homozgosity for the MCP-1 G allele at rs1024611 was associated with 4.7-fold greater risk of HAD. Further analysis found this allele to be associated with greater transcriptional activity, enhanced protein production, increased serum MCP-1 levels, and increased monocyte infiltration of tissues. |
| Singh et al. 2003 32 | 1049 HIV+ children; median age was 2.4 years; majority (59.7%) was non-Hispanic black. Participants followed for up to 36 months. |
Neurologic deterioration, a decline in neurocognitive test scores, and/or brain growth failure was considered evidence of disease progression between baseline and up to 36 months. |
Heterozygote carriers of the Delta 32 deletion in the CCR5 gene (rs333) exhibited delayed disease progression, lower frequency of cognitive impairment at baseline, and lower frequency of either impairment at baseline or a decline in neurocognitive status (trend level) when compared to homozygous wild-type carriers. Among homozygotic carriers of the Delta 32 deletion, the most rapid disease progression was associated with A/A genotype at rs1799987 (CCR5). A/A genotype at rs1801157 on the SDF-1 gene was also associated with faster disease progression, including neurocognitive impairment over time. This was relatively uncommon, occurring in <2% of children studied. Modest or little effects were documented for rs1799864 (CCR2), or two SNPs on CCR5 locally designated as 59356 and 59353. |
| Cutler et al. 2004 83 | 10 HIV+ African American males This was a postmortem study of brain tissue comparing sphingolipids and sterols in the medial frontal cortex, parietal cortex, and cerebellum of HIV-Dementia patients |
HIV Associated Dementia (determined by the presence of encephalitis in brain tissue and pre-mortem Memorial Sloan- Kettering scale > 1). |
The ApoE 4 allele was associated with dysregulated lipid and sterol metabolism, as well as elevations of sphingomyelin, ceramide, and cholesterol in the medial frontal cortex, parietal cortex, and cerebellum. The 4 allele was not related to astrocytes or activated microglia |
| Diaz-Arrastia et al. 2004 66 | 270 HIV+ persons who died from AIDS complications (unknown demographics); two separate cohorts assessed (one cohort whose members died during the monotherapy era; another whose members died during the dual therapy era). |
HIVE, as well as the presence of any of the following pathologies: microglial nodules, multinucleated giant cells, myelin pallor, and vacuolar myelopathy |
No association between pathological findings and the ApoE ε4 allele (rs429358 and rs7412), TNF-2, IL-1B*2, and ILIRN*2 |
| Singh et al. 2004 43 | 121 HIV+ cognitively normal participants; the majority were Caucasian, non-Hispanic (68%) and male (88%). Prospective study with a median follow-up of 3.9 years and cognitive retesting every 6-12 months. |
Neurocognitive impairment, defined as a Clinical Rating score of 5 or higher, and based on comprehensive neurocognitive testing234 |
At baseline, none of the allele examined (CCR5 delta-32 deletion; MCP-1-2518, CCR2-V64I) were associated with neurocognitive impairment rates. In the longitudinal analysis, possession of one or two CCR2-64I alleles at rs1799864 was associated with earlier progression to neurocognitive impairment from study entry or from estimated time of seroconversion. Null findings: Delta 32 deletion at rs333 (CCR5), and rs1024611 (MCP-1-2518-G/A) |
| Valcour et al. 2004 235 | 182 HIV+ adults (N = 85 <40 years; N = 97 ≥ 50 years); Sample was 54% Caucasian, 32% Asian or Pacific Islander, and 14% other, |
HAD (AAN, 1991), as determined via standardized neuropsychological testing, brain MRI, and serum tests |
A significant association was observed between the APOE ε4 allele (rs429358 and rs7412) and HAD among older (age ≥ 50 years) but not younger (<40 years) participants. |
| Shiramizu et al. 2006 236 | Repository CSF specimens from 27 prenatally- infected with HIV. |
HIV-Associated Encephalitis | MCP-1 2578G allele at rs1024611 was significantly more common in children with high HIV DNA in CSF. This allele was also associated with higher levels of supernatant MCP-1 in the CSF. |
| Burt et al. 2008 75 | 1,267 HIV+ adults; ethnically diverse (54% Caucasian); majority were male; they were followed prospectively with a median follow-up time of 5.9 years. |
HIV Associated Dementia. Criteria not specified; however, the cohort appears to be the same as Gonzalez et al (2002)39, in which criteria were derived from the Center for Disease Control233, and included neuropsychological testing and neuroimaging. |
ApoE ε4 allele was not associated with time to development of HAD, though it was associated with accelerated disease progression and time to death. |
| Pomara et al. 2008 71 | 41 non-demented HIV+ adults; predominantly African American (63%) and male (63%). This study assessed the effect of the ApoE ε4 allele on memory following acute Lorazepam administration |
Performance on verbal learning/memory and psychomotor tests |
The ApoE ε4 allele (rs429358 and rs7412) was associated with better immediate and delayed verbal recall at baseline assessment only. |
| Pemberton et al. 2008 70 | 56 Caucasian HIV+ adults with HAD/ADC, stage ≥ 1 and CD4 count < 500 cells/μL; other demographics unknown Data were also combined with genetic data from participants of other studies for a meta-analysis. This included HIV+ and HIV− controls. |
ADC/HAD, stage ≥ 1 (moderate to vegetative HAD) per 237 |
HAD was more common among individuals who were homozygous for A allele at rs1800629 (TNF-alpha-308). When this data was combined with previously published data, possession of just one A allele was associated with HAD. No differences in allele frequencies were found for rs3783525 (IL1A-889), IL1B+3953, IL12B 3′UTR. ApoE genotype (rs429358 and rs7412) did not differ between HAD patients and HIV+ controls in this study or when the data was combined with previously published studies. |
| Levine et al. 2009 40 | 143 HIV+ adults (primarily Caucasian and African American) who were either neurologically normal (N = 117) or who met criteria for HIV-associated dementia (N = 26) per established AAN criteria. |
HIV-associated dementia (HAD) per AAN criteria. Diagnosis was established via standardized neuropsychological testing and neuromedical exam. |
TT genotype at rs1130371within the CCL3 gene was associated with a two-fold greater risk of HAD. Depression was associated with a five-fold greater risk of HAD. There was no association between HAD and any other of the other polymorphisms studied: rs1024611 (MCP-1), rs1719130 (CCL5), rs17561 (IL-1a), rs1800872 (IL-10), rs1800629 (TNF- a), rs1801157 (SDF-1) |
| Bousman et al 2010 238 | 192 sexually active men with and without methamphetamine dependence (METH+/−) and/or HIV infection (HIV+/−). Ethnicity was 71% Caucasian, 15% African American, and 14% Hispanic |
Executive functioning domain Deficit Score |
There was a main effect of executive functioning but not of COMT Val158Met (rs4680) genotype on the total number of sexual partners. There was an interaction between rs4680 and executive functioning on total number of sexual partners and insertive anal sex (among Met/Met and Val/Met but not Val/Val carriers). |
| Joska et al. 2010 69 | 144 HIV+ young adults just entering care in South Africa, where Clade C is more common; the majority was female (74%) |
HAND based on Frascati criteria (Antinori et al. 2007) using standardized neuropsychological testing. |
Null findings between ApoE genotype and level of HAND severity. When comparing just HAD to no-HAD, the ε4 allele (rs429358 and rs7412) was less common in HAD. |
| Spector et al., 2010 72 | 201 Chinese HIV+ adults predominantly (93%) co- infected with Hepatitis C. |
Global Deficit Score based on standardized neuropsychological testing. Considered both cross- sectional comparisons and rtes of changes in neurocognitive status over the 12 months study period |
A higher percentage of ApoE 4 carriers (rs429358 and rs7412) were cognitively impaired at baseline. MBL-2 genotype (based on rs1800450, rs1800451, and rs5030737) was associated with neurocognitive changes over a 12-month period: 53% of those with O/O genotype declined whereas 23% of those with A/A genotype declined. No significant differences in baseline neurocognitive ability or change over time were observed for rs1799987 or rs333 (CCR5), CCR2-180-G/A, rs1801157(SDF- 1), IL4-589-C/T, rs1024611 (MCP-1), CX3CR1-745-G/A and - 849-C/T SNPs, or for the CCL3L1 copy number variant. |
| Sun et al. 2010 73 | 44 HIV+ male adults ; 11 HIV− adults; 62% Caucasian; all male. Ethnicity included 62% Caucasian and 22% African American) |
Neuropsychological impairment (>1.5 SD below normative mean in two domains on a comprehensive test battery |
ApoE genotype (rs429358 and rs7412) was not associated with neurocognitive outcomes |
| Andres et al. 2011 64 | 48 HIV+; 39 HIV-; Majority were male; largest ethnic groups were Caucasian (40%), Native or part-Native Hawaiians (28%), and Asians (13%) |
Global Cognitive Score, based on the average of several domain Z-scores from a comprehensive neurocognitive test battery). The HIV Dementia Scale239 was also examined as an outcome. |
Significant interactions were found between ApoE genotype and HIV serostatus, with HIV+ 4 allele carriers (rs429358 and rs7412) performing significantly worse than HIV seronegative ε4+ controls and seronegative ε4− controls on Global Cognitive Score and several specific cognitive domains. Among the HIV+ individuals, 4 carriers performed worse than non-carriers on the HIV Dementia Scale. |
| Chang et al. 2011 62 | 69 HIV+ adults of mixed ethnicity; predominantly male 70 HIV− adults of mixed ethnicity; predominantly male |
Domain Z-scores and a Global Z-score were determined based on a comprehensive neurocognitive test battery. |
The ApoE ε4 allele (rs429358 and rs7412) was associated with poorer neurocognitive functioning (verbal fluency, executive function, learning and memory) and smaller brain volume in HIV+ participants. This allele demonstrated a positive effect among HIV− individuals. Further analysis by age group (older vs. younger), HIV sero- status, and APOE genotype indicated that the 4 allele had a deleterious impact on younger HIV+ individuals and a positive effect among younger HIV− individuals. |
| Gupta et al. (2011)240 | 310 ethnically diverse males separated into four groups: 56 HIV-/methamphetamine nonusers, 77 HIV−/ methamphetamine users, 84 HIV+/ methamphetamine nonusers, 93 HIV+ methamphetamine users |
Neurocognitive impairment, defined by a Deficit Score cutoff of ≥0.5. Based on a battery of neuropsychological tests. |
The C allele at rs6280 of the Dopamine receptor 3 gene was associated with greater rates of neurocognitive impairment only among HIV+/methamphetamine users |
| Bol et al. 2012 41 | 86 HAD cases and 246 non-HAD AIDS patients as controls. All cases were from the Netherlands |
Diagnosis of HAD was determined in various ways due to the retrospective nature of the data. This included DSM, AAN, and Frascati criteria. |
The Delta-32 deletion in the CCR5 gene (rs333) was associated with HAD in cases who developed AIDS prior to 1991, but not after. Prep1 genotype (rs2839619) differed between cases and controls irrespective of year of AIDS diagnosis. Null findings for: rs429358 and rs7412 (ApoE); rs1130371 (CCL3); rs1799864 (CCR2); rs12483205 (DYRK1A); rs1024611 (MCP-1); rs1046099 (MOAP1); rs12909130 (PDE8A); rs17519417 (SPOCK3); rs1800629 (TNF-alpha); rs2905 (UBR7) |
| Brown et al. (2012) 46 | 262 HIV+ individuals; 60% African American; | HIV dementia severity as determined by the Memorial Sloan Kettering (MSK) classification. |
There were no differences in CCL3L1 copy number in relation to HIV dementia severity. |
| Levine et al. (2012)111 | 184 HIV+ adults (primarily Caucasian and African American). All cases were diagnosed as neurologically normal, or with mild cognitive/motor disorder or HIV-associated dementia per established AAN criteria, or subsyndromic HIV-related neurocognitive impairment equivalent to asymptomatic neurocognitive impairment as per 2007 Frascati criteria |
Neuropsychological domain T- scores (working memory, processing speed, learning, memory, motor). Scores were standardized based on entire NNTC cohort. |
Regression analysis found that COMT val158met (rs4680), BDNF val66met (rs6265), or DAT 3 (3′ UTR 40bp) genotypes did not predict neurocognitive functioning after controlling for disease severity, depression, and demographic variables. |
| Soontornniyomkj et al. (2012) |
Brain tissue from an ethnically diverse sample of 160 HIV+ and 22 HIV− adult persons. The majority were male. |
HAND and Aβ plaques. HAND (mild cognitive/motor disorder or HAD per AAN criteria or subsyndromic impairment equivalent to Asymptomatic Neurocognitive Impairment per 2007 Frascati criteria). Evaluation included comprehensive neurocognitive testing and neuromedical examination. |
ApoE ε4 allele (rs429358 and rs7412) and older age (≥50) were independently associated with increased likelihood of cerebral Aβ plaque deposition. Although the ApoE ε4 allele did not increase risk of HAND independently, ε4 carriers with Aβ plaque deposition had a higher risk of HAND. |
| Morgan et al 2013 241 | 466 HIV+ adults; 50% Caucasian; 78.8% male | HAND based on 2007 Frascati criteria |
This cross-sectional study found no effect of ApoE ε4 allele (rs429358 and rs7412) on HAND, or HAD in particular. No interaction between ApoE 4 allele and age, ethnicity, substance use disorders, duration of infection, or nadir CD4 on risk for HAND was observed |
| Panos et al. 2013 242 | 259 HIV+ ethnically diverse adults (55.2% Caucasian) |
HAND (mild cognitive/motor disorder or HAD per AAN criteria or subsyndromic HAND equivalent to asymptomatic neurocognitive impairment as per 2007 Frascati criteria). Evaluation included comprehensive neurocognitive testing and neuromedical examination. |
94% of older APOE 4 allele carriers had HAND compared to 56% non-carriers among older participants (age ≥ 50 years). No association between 4 and HAND was found for younger (< 50 years) participants. Analysis by cognitive domain revealed the combination of advanced age and the 4 allele was associated with poorer executive functioning and information processing speed. |
| Singh (2013)243 | 1,049 HIV+ symptomatic pediatric patients; predominantly of minority status (60% African American, 26% Hispanic). Participants were enrolled prior to combined antiretroviral therapy availability Longitudinal study with median follow-up time of 18.6 months |
CNS impairment defined as time from to deterioration in brain growth, psychological function, and/or neurological status. Neurocognitive decline was defined as the absence of any increase in raw scores or a decline in normalized scores by 2 SD for children <30 months of age, or by 1 SD for older children. Deterioration in neurologic function was defined as the loss of previously documented motor skills, reflexes, or behavior. |
APOBEC3G-H186R (rs8177832) G/G genotype was associated with CNS impairment compared with the wild-type A/A or A/G genotype. Both additive and dominant models found the APOBEC3G-F119F-C allele (rs5757465) to protect against CNS impairment. |
| Hoare et al. (2013)244 | 24 HIV+ individuals with at least one ApoE ε4 allele were compared to 19 HIV+ without ε4 allele. Participants were young, mostly female, and of Xhosa origin. This study was conducted in South Africa, where Clade C is more common; the majority was female (74%) |
Group comparisons in neuropsychological functioning and diffuser tensor imaging were conducted. |
The ε4 group had poorer immediate and delayed verbal memory and decreased fractional anisotrophy in the corpus callosum. |
| Genome Wide Association Studies | ||||
|---|---|---|---|---|
| Study 1st author (year) |
Sample Description | Genotyping Platform | Phenotype | Findings |
| Bol (2011)(36) | In vitro infection with HIV-1 of human monocyte- derived macrophages |
Illumina 610 Quad beadchip |
Production of p24 (low vs. high) |
No findings were significant at the GWA threshold. The strongest association was found for rs12483205 in the DYRK1A gene (p = 2.16 × 10−5). |
| Levine (2012)(37) | 1,287 individuals of Northern European ancestry |
Illumina 1M, 1MDuo, or 550K. |
Changes in executive functioning and processing speed between two visits up to 15 years apart; prevalence of HAD (AAN); and prevalence of neurocognitive impairment as determined by comprehensive neurocognitive testing every two years. |
No significant relationships were found between any of the over 2.5 million SNPs genotyped and phenotypes. SNPs within two genes, SLC8A1 and NALCN, had p-values of 3.31 × 10−7 and 6.62 × 10−7, respectively. Previously implicated SNPs, including rs1130371 (MIP1-a), rs1800629 (TNF-a), rs1024611 (MCP-1), rs1801157 (SDF-1), rs2839619 (Prep1), and MBL (rs1800450, rs1800451, and rs5030737) were not significant. |
When possible the Research SNP (rs) number is provided
2.1.1. Immune-related genes
There is a wide variety of immune factors that have been implicated in the chronic neuroinflammatory state leading to HAND11. These largely involved cytokines, chemokines, and their cell surface receptors. Both cytokines and chemokines can affect HAND neuropathogenesis via numerous routes. HIV requires chemokine co-receptors to enter cells12,13. As such, chemokines compete with HIV at these co-receptors and subsequently modify HIV replication14 and disease progression15,16. Chemokines also affect macrophage activation and chemotaxis of monocytes and other cells across the blood–brain barrier17,18, thus leading to increased inflammation and viral seeding of the CNS. Further, chemokines can affect neuronal signaling with subsequent disturbance of glial and neuronal functions 19,20.
Leveraging the increasing knowledge of how chemokines, cytokines, and other immune factors influence HAND pathogenesis, many groups have utilized candidate gene association studies to characterize how specific genetic susceptibility loci within immune-related genes modify risk for HAND 21-24. Here we briefly discuss some of these genetic susceptibility loci. Additional detailed information regarding their biological roles of these gene products can be found in25
C-C chemokine receptor type 5 (CCR5) is the most common HIV-1 co-receptor, at least during the early course of infection. CCR5 mediates gp120 neurotoxicity26. A 32-basepair deletion in the CCR5 gene, resulting in the CCR5-Δ-32 allele (rs333), leads to structural changes within the HIV co-receptor that confers high resistance to HIV infection among those who are homozygous 27,28. Early studies suggested that this allele conferred protection against HAND. For example, Boven and colleagues 29 found that not a single case among their sample of European American individuals diagnosed with HIV-associated dementia had a CCR5-delta-32 allele, which normally occurs in 10-20% of individuals with northern European ancestry. While this was soon confirmed by others30, more recent studies have not found an association31,32. Bol et al 33 observed that the delta-32 genotype was associated with HAD in individuals who developed AIDS prior to 1991, but not after, which was interpreted as reflecting the waning effect of this genotype on viral load set point. Still, looking at neurocognitive functioning rather than HAND diagnosis, Singh and colleagues 34 found that children heterozygous for the CCR5-Δ-32 allele had slower disease progression and less cognitive impairment than those homozygous for the wild-type.
Monocyte chemoattractant protein-1 (MCP-1, or CCL2) is a chemokine that recruits monocytes and other immune cells into the CNS, and is therefore believed to be responsible in part for the neuroinflammatory response. In vitro HIV infection of human leukocytes results in increased transmigration across the blood brain barrier (BBB) in response to MCP-1, and increased transmigration is correlated with increased expression of MCP-135. Elevated levels of MCP-1 have been detected in the brain and CSF of patients with HIVE and HAD 36,37, and are positively associated with dysfunctional CNS metabolism11. Further, the HIV protein Nef has been observed to induce MCP-1 expression in astrocytes with subsequent infiltration of infected monocytes into the brain 38. A single nucleotide polymorphism in the MCP-1 gene, resulting in the MCP-1-2578 allele, leads to increased levels of MCP-1 in serum 39 and CSF 40, and has been linked to accelerated disease progression and a 4.5 fold increased risk of severe HAND 41, although this finding has not been consistently replicated31,42. Another recent study found a significant difference in Prep1 allele distribution among HAD cases and non-HAD HIV+ controls33. Prep1 is a transcription factor with preferentially binding in the promoter region of the MCP-1 gene. In addition, a polymorphism within the minor HIV co-receptor CCR2, the natural target receptor for MCP-1, has also been connected to slower HIV disease progression 43. Individuals heterozygous for the CCR2-V64I allele exhibited slower disease progression and developed AIDS 2-4 years later than those who were homozygous for the wild-type allele. A later study found CCR2-V64I to be associated with slower progression towards neurocognitive impairment 32.
Macrophage inflammatory protein 1-alpha (MIP-1α, also known as CCL3) is a chemokine and natural ligand of the HIV co-receptor CCR5. MIP-1α expression is increased in the brains of those with HIVE, and released by both microglia and astrocytes 44. A SNP (rs1130371) within the MIP-1α gene was previously associated with HIV disease progression 45 and was found to be associated with a two-fold greater risk for HAD42 in the National NeuroAIDS Tissue Consortium cohort. More recently, our group has found an interactive effect between another SNP (rs1719134) and HIV status upon learning ability changes over time, such that HIV+ individuals show less improvement over multiple testings as compared to their HIV-negative counterparts, although the difference was small from a practical standpoint. These two markers (rs1130371 and rs1719134) are in high linkage disequilibrium, and the findings from this more recent analysis in the Multicenter AIDS Cohort Study cohort validate the role of MIP-1α in HAND.
Tumor necrosis factor-alpha (TNF-α) is an inflammatory cytokine produced by macrophages and microglia that is involved in apoptosis, viral replication, and in the regulation of immune cells 46,47. Increased levels of TNF-α mRNA have been found in macrophages derived from individuals with HIV-associated dementia48. Elevated TNF-α has a myriad of adverse effects upon the brain, including the potentiation of glutamate neurotoxicity 49, disruption of ionic transport in astrocytes 50, damaging of oligodendrocytes 51 and cortical neurons 52, and increasing the permeability of the blood brain barrier 53. A SNP within the promoter region of the TNF-α gene (rs1800629) is associated with response to viral proteins. Possession of even one allele containing this SNP was associated with HAD, as compared to HIV+ individuals without dementia and a healthy control population 54. This finding was recently replicated in meta-analysis21, but was not confirmed in another study 42.
Stromal cell-derived factor-1 (SDF-1; also called CXC Chemokine Ligand 12, or CXCL12), a major ligand for the major HIV co-receptor CXCR4, has been noted to inhibit HIV-1 transmission by competing for CXCR4 binding due to its high expression in genital and rectal epithelium 55. In addition, SDF-1 down-regulates expression of CXCR4, hindering infection by T-tropic HIV-1 strains. In the context of HAND, however, mRNA levels of SDF-1 are elevated in HIVE when compared to uninfected controls 56,57, suggesting that once infected, this chemokine is associated with the pathogenesis of HAND. This hypothesis is bolstered by findings from in vitro studies demonstrating that SDF-1 is toxic to neurons 57-59. A SNP in the SDF-1 gene, resulting in the SDF1-3′-A allele (rs1801157), has been found to delay the onset of AIDS 60. Conversely, Singh and colleagues 34 reported that African American children that were homozygous for SDF1-3′-A had more rapid progression and decline in neurocognitive ability, but noted that this genotype was very rare in their sample. It may be that chemokines such as SDF-1 play different roles throughout development, explaining the contradictory results between studies using adults and those using children. This, as well as ApoE discussed next, may be examples of antagonistic pleiotropy in the context of HAND61.
-
Apolipoprotein E (ApoE) is primarily involved in catabolism of triglyceride-rich lipoproteins, but also mediates innate immune response including macrophage immune responsiveness 62. The ApoE ε4 allele has long been implicated in the pathogenesis of Alzheimer’s disease, and more recently has become of interest in the pathogenesis of HAND 61,63-73. Among the earliest studies, Corder and colleagues 67 found twice as many individuals carrying at least one ε4 allele were given a diagnosis of dementia (specific criteria were not specified) over the course of the five-year study. Subsequent studies over the last decade, however, have yielded inconsistent findings. Potential mitigating factors include the deleterious influence of the ε4 allele on disease progression and survival rates 74-76, methodological differences between studies in the operationalization of HAND, and differences between studies in terms of the inclusion of a HIV seronegative control sample. Recent studies using such a control sample and objective measures of neurocognitive functioning suggests synergistic deleterious effects of the ε4 allele and HIV on cognition 61,63. Within HIV+ samples, age has been found to be a modulating factor in some studies 64,73 but not others 61,71,77. Valcour and colleagues 73, for example, found that older ε4 carriers (age ≥ 50 years) had higher rates of HAD compared to age-matched ε4 non-carriers. Among their younger sample (<40 years of age), however, the proportion of individuals with HAD was comparable between ε4 carriers and non-carriers. Using the broader criteria of HAND as the outcome variable, similar findings were recently documented in our laboratory 64. Of the published longitudinal studies of ApoE genotype and HAND 67,71,74 none have employed a design aimed at measuring individual changes on objective neurocognitive measures over time, while also accounting for mitigating factors such as disease severity. Such an approach may help clarify the nature of the relationship between ApoE genotype and HAND.
Probing neuropathological correlates of ApoE, Diaz-Arrastia and colleagues 78 and Dunlop and colleagues 79 did not find a consistent association between ε4 and pathologic findings of HIVE or HAND, although they suggested that lack of sufficient statistical power may have resulted in the null findings. In addition, while HIVE is thought to be a common pathological substrate for HAD, the two can occur independently of one another 80. It should also be noted that some of these studies were conducted with patients who died in the pre-HAART era. Given the deleterious relationship between the ε4 allele and disease severity, individuals may have died before pathological effects emerged in the brain. ApoE-ε4 is associated with faster disease progression, possibly due to enhanced HIV fusion/cell entry 81. In contrast, Cutler and colleagues 82 found evidence for lipid metabolism dysfunctions in the brain tissue of ε4 carriers. Their sample also consisted of patients who died in the pre-HAART era. More recently, Soontornniyomkij et al. 66found that ε4 and older age were independently associated with increased the likelihood of cerebral Aβ plaque deposition in HIV+ adults. While the Aβ plaques in HIV brains were immunohistologically different from those in brains of individuals who died with symptomatic Alzheimer’s disease, this study is among the first to provide a link between genotype and neuropathological findings in HIV. As discussed further below, this new focus on neuropathological markers may prove more fruitful in dissecting the influence of host genotype upon risk for HAND.
Mannose-binding lectin-2 (MBL-2) is involved in the body’s innate immune response 83 and low concentrations are associated with greater susceptibility for infection and faster disease progression84. A collection of variants within the MBL-2 gene at rs5030737, rs1800450, and/or rs1800451 (referred to as the D, B, and C alleles, respectively) were found to be associated with cognitive decline among a Chinese cohort 22. Specifically, within a cohort of Chinese men, a greater percentage of individuals who were homozygous for any of these variants demonstrated neurocognitive decline after one year as compared to individuals who did not have any of these mutations. This same genotype was associated with more rapid decline towards neurocognitive impairment among a pediatric cohort85. Interestingly, MBL-2 is over-expressed in the neuronal axons of individuals with HIVE, and expression is positively correlated with MCP-1 expression in these individuals86.
2.1.2. Catecholamine related genes
In recent years, there have been numerous reports of polymorphisms within catecholamine, and more specifically dopamine (DA)-related genes, resulting in measureable differences in neurophysiological and neurocognitive functioning in non-HIV cohorts. Among the most commonly examined are the catechol-O-methyl-transferase (COMT) val158met allele87-95, the dopamine transporter-1 (DAT-1) 3′-UTR variable tandem repeat 96-101, and the brain derived neurotrophic factor (BDNF) val66met allele 102-109. While the effects of these variants upon neurocognitive phenotypes has been small, it is conceivable that among vulnerable individuals in whom DA functioning is already compromised, such as those with HIV99,110-113, the effects will be additive or synergistic. Thus far, cross-sectional studies have not indicated that these alleles increase one’s risk of HIV-related neurocognitive deficits114. For example, Levine et al114, examining cross-sectional data from the National NeuroAIDS Tissue Consortium, did not detect any interactive effect of disease severity (as measured by CD4+ T-cell count) and COMT, DAT1, or BDNF genotypes described above upon a number of neurocognitive domains in an exclusively HIV+ sample. Bousman et al. (2010) reported interactive effects of COMT val158met genotype and executive functioning on sexual risk taking in both HIV+ and HIV- individuals 115. While no differences in executive functioning were noted between groups, they did find that among Met allele carriers, those with greater executive functioning deficits reported greater number of sexual partners and other risky sexual practices.
The additive or synergistic effects of DA-related alleles and stimulants such as methamphetamine and cocaine in HIV+ cohorts have also been examined. Gupta et al 116 investigated the impact of a SNP (rs6280) within the dopamine receptor-3 (DRD3) gene upon neurocognitive functioning in four groups, stratified for HIV status and methamphetamine use. The biological connection between DRD3 and HAND is especially interesting, as macrophages are increasingly infected by HIV in the presence of both methamphetamine and increased extracellular DA individually, and that this process is mediated by DA receptors expressed in macrophages, including as DRD3. As the authors hypothesized, only the HIV+ methamphetamine users were found to have genotype-related neurocognitive alterations.
Analyzing longitudinal neurocognitive data from the Multicenter AIDS Cohort, our group has recently examined the longitudinal interaction between HIV status, stimulant use, and COMT genotype in a very large cohort (N = 952) that included both HIV+ and HIV-seronegative individuals. COMT genotype was found to influence the longitudinal neurocognitive functioning of only HIV-seronegative individuals117. Further, DA-related genetic variants in BDNF (rs6265), dopamine-β-hydroxylase (rs1611115), DR2/ANKK1 (rs1800497), and DR3 (rs6280) did not impact the longitudinal neurocognitive functioning of HIV+ individuals, despite the well documented involvement of DA functioning in HAND.
2.2. Genome-wide association studies
Genome-wide association studies (GWAS) have become increasingly affordable and a practical means to study disease pathogenesis. Several such studies have identified additional risk variants associated with HIV disease progression, viral set point, rapid progressors, and other disease-related phenotypes, as previously reviewed 118,119. GWAS have also recently proven valuable for the study of already relatively well-characterized neurologic diseases. As an example, a recent GWAS of late-onset Alzheimer’s disease resulted in the identification of several new genetic susceptibility loci 120-123. With regards to HAND, for which the neuropathogenesis is less understood, GWAS may hold even greater promise. This is because it is unlikely that one or a few genetic susceptibility loci confer a large proportion of the risk for HAND. Instead, variability in HAND risk may depend on a range of common variants, which are more amenable to detection via GWAS platforms (yet require very large sample sizes). As such, the genetic variants detected by GWAS may help to achieve an improved mechanistic understanding of the disease and ultimately lead to targets for pharmaceutical treatment and prevention 124. Only one GWAS focusing on HAND has been published to date125. The study included 1287 Caucasian adults enrolled in the Multicenter AIDS Cohort Study (MACS). Leveraging the MACS longitudinal protocol that includes serial neurocognitive testing and neuromedical examinations, as well as prevalence of HAD, this study examined a variety of neurocognitive phenotypes for association with over 2.5 million SNPs. No genetic susceptibility loci were associated with the phenotypes, which included decline in processing speed or executive functioning over time, prevalence of HAD, and prevalence of neurocognitive impairment based on a comprehensive neuropsychological battery. Two SNPs, within the SLC8A1 and NALCN genes, had p-values just below the strict GWAS threshold in association with change in processing speed over time. SLC8A1 (solute carrier family 8 [sodium/calcium exchanger] member 1) is involved in ion channel/ion transporter activity in plasma and mitochondrial membranes. Both SLC8A1 and NALCN (sodium leak channel, nonselective) are involved in sodium transport across cellular and intracellular membranes. Loss of mitochondrial membrane potential and other effects of ion transport dysfunction have been implicated in HAD 126-128. Due to the relatively small sample size, future collaborative efforts that incorporate this dataset may still yield important findings.
2.3. Summary
The value of targeted candidate gene association studies for investigating HAND neuropathogenesis is that HAND is a syndrome that is remote from its cellular causes. However, a requisite for such studies is that the genes under investigation meet a standard of biological plausibility. Accordingly, candidate gene studies have implicated a variety of immune-related gene variants as risk or protective factors for HAND; however, very few have been replicated in later studies. One reason for this, as indicated in Table 1 and discussed further below, is the lack of a reliable and consistently applied phenotype for HAND. Another reason is that thus far the focus has been SNPs, tandem repeats, and microdeletions. Other types of genetic variation, such as copy number variants, have not been thoroughly examined in this context. Finally, significant associations may often be spurious, due in part to population stratification and admixture, not to mention lack of statistical power. Therefore, recruitment methods and statistical strategies must be especially rigorous. With regards to GWAS, collaborations across cohorts with the goal of increasing the statistical power to detect common variants contributing to neuropathogenesis will be necessary, and supplemental strategies to follow-up GWAS analysis may also be useful for revealing associations left undetected initially129.
On a more sobering note, recent findings from our group suggest limited utility of genetic studies for elucidating the neuropathogenesis of HAND. Preliminary findings from a study of MACS-based study that examined the longitudinal neurocognitive data from 952 HIV+ and HIV-seronegative individuals did reveal that MIP-1α (CCL3) and MCP-1 (CCL2) genotype did affect neurocognitive functioning over time as a function of serostatus; however, the effect is very minor. As such, while these findings may further underscore the importance of these immune factors in HAND neuropathogenesis, the limited effects of individual gene variants on HAND may not particularly useful for informing treatment strategies or further clarifying the pathogenic pathways that lead to HAND. As such, greater focus might be placed on dynamic cellular processes, described next.
3. Transcriptomic studies of HAND
3.1. Brain-Based Gene Expression Studies
Recent gene expression studies have taken advantage of genome-wide microarrays, allowing surveillance of virtually the entire transcriptome. Coupled with bioinformatics and statistical methods coming from the field of systems biology, putative biological networks associated with disease state can be identified 130,131. Gene expression studies have been widely used to investigate and discover cellular mechanism involved in the pathogenesis of HIV (as reviewed in 132). In the context of HAND, there have been a fast growing number of global transcriptome studies to date. Some investigators have focused on specific brain cells in vitro 130,133-135, using such as methods such as laser capture microdissection. However, with the availability of affordable microarrays, most transcriptome studies to date have utilized brains of HIV-infected (HIV+) humans, and have focused on gene expression changes underlying HIVE. Such studies, generally limited to examination of homogenized frontal grey matter tissue, have found altered regulation of genes involved in neuroimmune functioning, and also implicated neurodegenerative pathways based on dysregulation of genes involved in synapto-dendritic functioning and integrity136, toll-like receptors137, and interferon response138. As recently reviewed139, findings from human microarray studies have been partially replicated in simian immunodeficiency virus (SIV) models, especially with regards to interferon-related and neuroinflammatory-related genes 140,141. Mouse astrocytes exposed to HIV have also shown some transcription overlap with those of simian and human studies142. Similarities among animal and human brain transcriptome studies were recently reviewed 143.
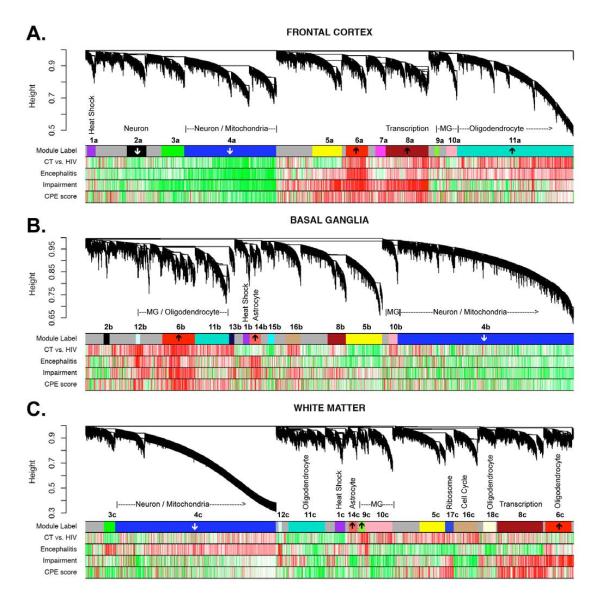
More recently, in analyzing gene microarray data derived from multiple brain regions of HIV+ individuals with HAND alone or HAND with HIVE, Gelman et al found two apparently distinct transcriptome profiles implicating two distinct etiological pathways to HAND144. HIVE with concomitant HAND was associated with high viral load (mRNA) in the brain, upregulation of inflammatory pathways across all brain regions, and downregulation of neuronal transcripts in frontal neocortex, whereas HAND without HIVE was characterized by low brain viral burden without evidence of increased inflammatory response, and without downregulation of transcripts in frontal neocortical neurons. Indeed, only transcripts characteristically expressed by vascular and perivascular-type cells were consistently dysregulated in HAND without HIVE. These data were recently re-examined by Levine et al145 using weighted gene coexpression network analysis (WGCNA)146. While standard gene expression studies such as Gelman et al144 utilize a group comparison approach, WGCNA enables a more systematic and global interpretation of gene expression data by examining correlations across all microarray probes, identifying biologically meaningful ‘modules’ that are comprised of functionally related genes and/or correspond to cell types147. WGCNA typically results in fewer than 20 modules (as opposed to thousands of genes), which can then be examined for their association to clinical or biological variables of interest. In Levine et al145, a number of biologically-meaningful gene expression modules were identified and then correlated with a global neuropsychological functioning index and CNS penetration effectiveness (Figure 1). While the WGCNA largely validated the findings from Gelman et al, it also identified meta-networks composed of multiple gene ontology categories, as well as oligodendrocyte and mitochondrial functioning.
Figure 1.
Co-expressed genes tend to cluster into modules corresponding to major biological divisions (cell type, molecular function, etc), as well as disease-relevant changes. Module summaries are presented for frontal cortex (top), neostriautm (middle), and white matter (bottom). The dendrograms group genes into distinct modules using hierarchical clustering. The y-axis corresponds to distance determined by the extent of topological overlap (1-TO). Top color band (Module label): dynamic tree cutting was used to identify highly parsimonious module definitions, generally dividing modules at significant branch points in the dendrogram. Second color band (CT vs. HIV): plotted T values of control (group A) vs. HIV+ samples (group B,C,D). Third color band (Encephalitis): plotted T values of HIV without encephalitis (groups B,C) vs. HIV with encephalitis (group D) samples. Fourth color band (Impairment): plotted correlations of GCR scores, our measure of neurocognitive impairment, across all HIV subjects. Fifth color band (CPE score): plotted correlations of CPE scores across all HIV+ subjects. NOTE: Red corresponds to genes with higher expression in HIV, impairment score (GCR), HIVE, and CPE score for color bands two through five, respectively. Note that the most significant correlations with disease tend to occur in FC. Arrows within top color band indicate that these modules are enriched for genes showing increased or decreased expression in hippocampus of Alzheimer’s disease (Blalock et al. 2004; p<0.00002). Figure from Levine et al 145.
Levine et al 145 also identified genes that were commonly associated with neurocognitive impairment in Alzheimer’s disease and HIV (Table 2). Specifically, common gene networks dysregulated in both conditions included mitochondrial genes, whereas upregulation of various cancer-related genes was found. Importantly, a recently published meta-analysis by Borjabad and Volsky (2012) compared global transcriptomes derived from frontal grey and/or frontal white matter from individuals with HIVE and/or HAND to those from individuals with AD (various anatomic locations), without consideration of NCI148. Both diseases (as well as multiple sclerosis) were associated with up-regulation of a wide range of immune response genes, and HAND and AD also shared down-modulation of synaptic transmission and cell-cell signaling. However, due to the different methodologies used, comparison to the Levine et al study 145 is not possible.
Table 2.
Enriched GO categories for common impairment genes. All genes in this list were correlated with the Mini Mental Status Exam in Blalock et al.245, as well as with Global Clinical Rating in the current analysis of HIV brains (both Frontal Cortex and Basal Ganglia, R>0.25).
| Genes down-regulated with impairment in both AD and HIV | ||||||
|---|---|---|---|---|---|---|
| Gene Category | List Hits |
List Total |
Pop Hits |
Pop Total |
EASE score | Bonferroni |
| Cytoplasm | 175 | 259 | 2277 | 4968 | 5.62E-13 | 1.40E-09 |
| Energy pathways | 26 | 259 | 128 | 5016 | 4.16E-09 | 1.04E-05 |
| Mitochondrion | 48 | 259 | 396 | 4968 | 3.27E-08 | 8.15E-05 |
| Tricarboxylic acid cycle |
10 | 259 | 20 | 5016 | 2.24E-07 | 5.58E-04 |
| Transit peptide | 28 | 217 | 184 | 4007 | 1.14E-06 | 2.85E-03 |
| Synaptic vesicle | 10 | 259 | 30 | 4968 | 1.31E-05 | 3.26E-02 |
| Genes up-regulated with impairment in both AD and HIV | ||||||
| Cell differentiation | 10 | 131 | 90 | 5016 | 4.79E-13 | 1.40E-09 |
| Activator | 11 | 105 | 131 | 4007 | 1.89E-03 | 1.00E+00 |
| Repeat | 42 | 105 | 1051 | 4007 | 1.98E-03 | 1.00E+00 |
| Cell communication | 50 | 131 | 1363 | 5016 | 5.20E-03 | 1.00E+00 |
| Regulation of transcription |
32 | 131 | 762 | 5016 | 5.70E-03 | 1.00E+00 |
| Phosphorylation | 26 | 105 | 582 | 4007 | 6.18E-03 | 1.00E+00 |
Another area in which global transcriptome studies have been used in the context of HAND is to understand the effects of cART. Borjabad et al were the first to examine the relationship between cART use and global brain gene expression 149. Notably, cART-treated cases were found to have transcriptome signatures that more closely resembled those of HIV-seronegative cases. Further, individuals who were taking cART at the time of death had 83-93% fewer dysregulated genes compared to untreated individuals. Despite this, in both treated and untreated HIV+ brains there were approximately 100 dysregulated genes related to immune functioning, interferon response, cell cycle, and myelin pathways. Interestingly, gene expression in the HIV+ brains did not correlate with brain viral burden, suggesting that even high CNS penetration effectiveness (CPE)150, which has been shown to reduce CSF viral load151, may not reduce transcriptomic dysregulation. Indeed, the absence of an association between CPE and brain transcriptome by our group when utilizing both standard differential expression analysis and WGCNA145 would help to explain the equivocal results of studies examining the relationship between CPE and HIV-related neurocognitive dysfunction to date 152-156.
3.2. Monocytes Gene Expression Studies
While the examination of brain tissue transcriptome informs our understanding of the mechanistic underpinnings of HAND, the peripheral blood system has been the focus in the search for biomarkers and upstream mechanisms. Only recently has global transcription analysis been used in this context. Like brain tissue, the first hurdle is to separate the desired blood cells before extracting mRNA for microarray analysis. Thus far, monocytes have been the cells of choice for blood transcriptome studies of HAND, and for clear reasons. CD16+ monocytes are among the first cells to become infected with HIV, and a subset of these cells also appears to be associated with the pathogenesis of HAND157,158. As the virus gains momentum and the immune system weakens, infected monocytes cross the blood brain barrier as “Trojan Horse” cells with increasing frequency, driven by both increased chemokine release in the CNS and the peripheral immune response 159-161. This increased traffic of monocytes into the CNS, which also included uninfected monocytes, further increases the expression of chemokines, leading to recruitment of even more monocytes in a feed-forward manner 162,163. Once inside the brain, monocytes differentiate into perivascular macrophages 164, where their role in the neuropathogenesis of HAND has been well-described and includes the release of pro-inflammatory cytokines, chemokines, interferons, and viral proteins that are harmful to nearby neurons and other cells 161,165-169. Thus, monocytes are activated to migrate to the CNS by two mechanisms; peripheral immune activation and a chemokine gradient released from cells within the CNS, respectively termed the “push” and “pull” of inflammatory cell recruitment161. This interaction between the CNS and circulating blood monocytes is a key mechanism underlying HAND, and as such, delineating cellular alterations that occur within monocytes during this process holds promise for identifying biomarker and pharmaceutical targets.
Buckner et al (2011) examined dynamic transcription changes in an in vitro model170. Starting with monocytes from healthy donors and infecting the cells with HIV, they created a CD14+CD16+CD11b+Mac387+ monocyte subpopulation in vitro, and found this phenotype to be especially capable of crossing a laboratory model of the blood brain barrier. Gene expression analysis revealed upregulation of chemotactic and metastasis-related genes, but not inflammatory genes. In addition, they described dynamic changes as the monocytes matured into macrophages, including an increase in the expression of enolase 2, followed by a decrease once the cell was fully differentiated. Osteopontin was also observed to have increased expression in the maturing monocytes. This important study demonstrated that the dynamic changes in monocyte transcription may provide clues about biological processes necessary for neuropathogenesis.
Pulliam et al (2004) isolated CD14+ monocytes from 23 HIV+ individuals with high or low viral loads, as well as HIV-seronegative controls, and used microarrays to characterize differentially regulated genes in the cells171. Monocytes from individuals with high viral loads (>10,000 copies) showed increased expression of CD16, CCR5, MCP-1, and sialoadhesin, suggesting an inflammatory phenotype. However, they noted that the monocytes are not activated in the “classical sense”; that is, they did not produce a number of proinflammatory cytokines common in the pre-cART era (e.g., IL-6, TNF-alpha). Further, they showed a transcription profile consistent with chemotactic properties. Specifically, among individuals with high viral loads, they found evidence of increased propensity for chemotaxis characterized by increase CCR5 and MCP-1. The authors concluded that the circulating monocyte has evolved in the cART era to have a chronic inflammatory state with additional chemotactic features, essentially a mature monocyte/macrophage hybrid.
Sun et al (2010) reported the first study in which blood monocyte global transcription was compared to neurocognitive functioning in HIV+ individuals. They examined whether or not monocyte gene expression and other peripheral factors (CD4, ApoE genotype, viral load, lipopolysaccharide and soluble CD14) were associated with neurocognitive impairment in a group of 44 HIV+ individuals on cART, as well as 11 HIV− seronegative controls172. The authors found that monocyte gene expression, which showed a chronic inflammatory profile in the HIV+ participants with high viral load, was not correlated with neurocognitive impairment. Further, none of the blood markers was associated with overall neurocognitive impairment.
More recently, the same group focused their analysis on neurophysiological measures rather than HAND173. Specifically, they examined whether peripheral immune activation was associated with brain metabolite concentrations, as measured by MRS. Thirty-five HIV+ on cART and 8 HIV-seronegative adults were examined. Approximately half of the HIV+ sample was considered to have mild neurocognitive impairment based on standardized testing (the diagnosis of HAND was not determined). Absolute concentrations of brain metabolites in the frontal white matter, anterior cingulate cortex, and basal ganglia were determined and then examined in relation to monocyte gene transcription and a global neurocognitive measure. Among the HIV+ participants, they found an interferon-alpha induced activation transcriptome phenotype that was strongly correlated with N-acetylaspartate in the frontal white matter. Notably, Interferon-gamma inducible protein-10 (IP-10) was strongly correlated with plasma protein levels, and plasma IP-10 was inversely correlated with N-acetylaspartate in the anterior cingulated cortex. This study is particularly remarkable as it is the first to connect transcription changes with putative HIV-related neurophysiological changes. As discussed below, we believe that this tactic holds the greatest promise for elucidating the neuropathogenesis of HAND.
Finally, our lab has recently utilized the Illumina HT-12 v1 Expression BeadChip to analyze monocyte-derived transcriptome data from 86 HIV+ individuals enrolled in the Multicenter AIDS Cohort Trial174. In contrast to the studies described above, which employed standard fold-change comparisons between the HIV+ and control group, we utilized WGCNA with an all HIV+ sample, as described above 146,175. Unlike Sun et al. 172, our standard differential expression analysis identified a number of individual gene transcripts that were significantly correlated with global neurocognitive functioning, after correcting for multiple comparisons. Of the 16 genes identified, many are involved in neuroprotection or neurodegenerative processes, including the interleukin-6 receptor, casein kinase 1-alpha-1, hypoxia up-regulated-1, low density lipoprotein receptor-related protein-12, kelch-like ECH-Associated protein-1 (KEAP-1) 176-191. Correlations between these gene transcripts and global neurocognitive functioning were in the expected direction considering their biological roles. The KEAP-1 findings are especially interesting, as they support a recently described role for nuclear factor E2-related factor 2 (nrf-2) in HAND192. Briefly, KEAP-1 sequesters nrf-2 in the cytoplasm193. Inhibiting the action of KEAP-1 allows more nrf-2 to enter the nucleus, where it promotes the expression of numerous anti-inflammatory and anti-oxidant proteins194. The exploration in recent years by neuroAIDS researchers of factors that modify the activity of nrf-2 (e.g., GSK3-β inhibitors195 and curcumin196) further points to the relevance of the KEAP-1/nrf-2 mechanism in HAND neuropathogenesis. As such, this pathway deserves further attention as a potential pharmacological target during early signs of HAND, or even as a prophylactic. In addition, the WGCNA identified two modules associated with global neurocognitive functioning. Gene ontology analysis of the significant modules indicates that mitotic cell cycle was positively correlated with global neurocognitive functioning, whereas translational elongation was negatively correlated.
3.3. Summary
Genome-wide transcriptome studies have implicated numerous genes and biological pathways in the neuropathogenesis of HAND. Human studies have been partially replicated in simian and murine models. One limitation of previous studies is the use of homogenized brain tissue, which contains mRNA from numerous cell types 136,140,197,198, thus making it difficult to determine cell-specific molecular processes. In addition, most studies describe gene expression from one brain region (e.g., frontal lobe), and those regional disease-related transcription changes may not reflect the disease-related transcription changes occurring in brain regions also commonly implicated in HAND (e.g., basal ganglia). Also, most in vivo studies utilizing brain tissue have sought to understand alterations in gene expression in brain tissue of humans or animals that expired in an advanced state of disease (i.e., HIV encephalitis or HIV-associated dementia). As such, it is uncertain if the findings of these studies will generalize to HAND in the current era. In tandem with studies of brain tissue have been investigations of monocyte transcriptome, which may provide clues about earlier stages of pathogenesis. The interpretation of transcriptome data utilizing systems biological methods open the way to novel therapeutic targets. For a more comprehensive discussion of both animal and human brain transcriptome studies, the reader is referred to an review by Winkler et al 143
4. Epigenetic studies of HAND
4.1. MicroRNA Studies
MicroRNAs (miRNAs) are small RNA molecules that modify transcription and translation via interactions with mRNA. Within the CNS, miRNAs regulate a variety of processes. As such, their role in neurologic disease has received increasing attention. Still, only a handful of studies examining the role of miRNA in HAND have been published. The first study examined the impact of Tat upon expression of selected miRNAs in primary cortical neurons in vitro 199. Tat was found to upregulate mir-128a, which in turn inhibited expression of SNAP25, a pre-synaptic protein. A second study involved examination of brain tissue; caudate and hippocampus from rhesus macaques with (4) and without (4) SIV-encephalitis (SIVE), and caudate from humans HIV-negative controls (6) and those with both HAND and HIVE (5, although only 3 were used for the microarray analysis)200. While three miRNAs were found to be elevated in SIVE and HIVE (miR-142-5p, miR-142-3p, and miR-21), the study focused on miR-21. This miRNA, largely known for its link to oncogenesis, was significantly upregulated in the brains of both HIVE and SIVE, and was specifically found in neurons. Further analysis revealed that it was also induced stimulation of N-methyl-D-aspartic acid receptors, which lead to subsequent electrophysiological abnormalities. Using a variety of experiments, the authors showed that miR-21 targets the mRNA of myocyte enhancer factor 2C (MEF2C), a transcription factor crucial for neuronal function and a target of miR-21, ultimately reducing expression of this mRNA. Immunohistochemistry examination supported this by showing diminished expression of MEF2C in neurons of HIVE and SIVE brains. Noorbakhsh et al conducted miRNA profiling in the frontal lobe white matter of 4 HIV-negative and 4 HIVE cases matched by age and sex126. Differential expression of multiple miRNAs was found. The authors used a two-fold cutoff as a decision point for further analysis, a common cutoff in gene expression studies. The authors also employed bioinformatics in their analysis. This included predicting the mRNA targets for each of the differentially expression miRNAs. Gene ontology term analysis was then performed for predicted mRNA targets to determine their functional classes, which revealed that most of the up-regulated miRNAs targeting genes involved in immune response and inflammation, followed by nucleotide metabolism and cell cycle. Perhaps paradoxically, considering findings from many transcriptome studies, inflammation-related genes also ranked first among the targets of down-regulated miRNAs. Cell death-related genes also ranked highly among targets of down-regulated miRNAs. Further, miRNAs targeting caspase-6 were downregulated, thereby allowing greater expression of this gene, which was confirmed with immunohistochemistry analysis in the astrocytes of HIVE brain sections.
Tatro et al utilized both global mRNA and miRNA expression analysis in an ambitious study with the goals of identifying changes in miRNA expression in the frontal cortex of HIV+ individuals, determining whether miRNA expression profiles could differentiate HIV from HIV with concurrent major depressive disorder (MDD), and developing a method for integrating gene expression and miRNA expression data201.Their sample consisted of HIV-negative controls, HIV+, and HIV+ with concurrent MDD. miRNA from 3 individuals within each group were pooled and used for the miRNA profiling, and mRNA for 3 individuals from both HIV+ groups were used for non-pooled mRNA profiling. Importantly, neurocognitive functioning was not considering in this study, ages varied widely between groups, and one of the HIV/MDD brains had pathology consistent with HIVE. With these caveats in mind, in HIV+/MDD group, more miRNAs were downregulated than in the HIV+ group, and miRNAs also tended to be more clustered around the same chromosomal regions. After identifying mRNAs that were significantly differentiated in the HIV/MDD group, and then identifying miRNAs that were dysregulated by at least 2-fold relative to the HIV only group, the authors utilized a Target Bias Analysis to determine the relationship between miRNA dysregulation and target-gene dysregulation. Using this method, they identified miRNAs belonging to four categories: 1) those with many dysregulated mRNA targets but of marginal statistical significance, 2) those with fewer dysregulated target-genes but with high statistical significance, and 3) those with numerous dysregulated gene targets that were of high statistical significance, and 4) those that did not have a significant number of dysregulated targets. The authors also identified a small number of genes with 39UTR target sequences compatible with unusually high number of miRNA. These were considered to be “hubs” for miRNA activity, and the authors outlined their biological roles and association with neuropsychiatric illnesses.
Finally, the most recent study of miRNA in the context of HAND neuropathogenesis examined the impact of viral protein R (Vpr) in a human neuronal cell line. More specifically, in order to investigate the mechanisms underlying the altered expression of cytokines and inflammatory proteins in CNS cells resulting from HIV infection, the authors performed both miRNA and gene expression assays using human neurons (primary cultures or cell line) treated with recombinant Vpr proteins. Vpr was found to deregulate several miRNAs and their respective mRNAs 202. As one potential mechanism for neuronal dysfunction, they found that expression of both miR-34a and one of its target genes (CREB) were dysregulated in the presence of Vpr. This study was the first to demonstrate a miRNA-dependent pathway through which Vpr damages neurons.
4.2. Histone Modification Studies
Chromatin structure, and therefore gene expression, can be modified by the acetylation and deacetylation of histone proteins, a process that is mediated by histone deacetylases (HDACs)203. HDAC inhibitors have been shown to improve cognitive ability and may be candidates for treating a variety of neurologic diseases 204,205. We are aware of only one study examining histone modification in the context of HAND neuropathogenesis. Saiyed et al examined the influence of Tat upon expression of HDAC2 in neuronal cells in vitro, and the subsequent effect of HDAC2 modification on regulating genes involved in synaptic plasticity and neuronal function206. HDAC2 expression was negatively correlated with expression of CREB and CaMKIIa genes, which are involved in neuronal regulation.
4.3. DNA Methylation Studies
We are aware of only one study utilizing DNA methylation in the context of HAND. Perez-Santiago presented early findings of a methylation study at the 19th Conference on Retroviruses and Opportunistic Infections in 2012207. The investigators hypothesized that levels of DNA methylation levels could predict neurocognitive decline. Seventeen HIV+ adults were examined at two time points. Genomic DNA was assayed with the Illumina Hi-Seq 2000. The phenotype consisted of change in mean neuropsychological scaled score (with correction for practice effects). This was then correlated with methylation profile at time point 1, which revealed 26 strongly positive correlated autosomal sites, and 18 negative correlated sites. Correlation between change in neuropsychological scaled score and methylation profile at time point 2 revealed 48 highly correlated autosomal sites, and 26 negatively correlated. The authors posited that positive correlations indicated methylation leads to improvement in neuropsychological functioning NP whereas a negative correlation indicated that de-methylation was associated with neurocognitive improvement.
4.4. Summary
Epigenetic studies of HAND neuropathogenesis are relatively recent, with most studies focusing on miRNA pathways in infected tissue or cells. A variety of miRNAs have been implicated, lending validation to previously identified neuropathogenic mechanisms, such as increased capsase-6 and mitochondrial dysfunction. CREB has been implicated in both miRNA and histone studies. Finally, one group reported an association between improved neuropsychological performance and global DNA de-methylation, although follow-up studies are needed to validate these findings.
5. Current challenges and proposed solutions
5.1. HAND phenotypes: Problems and Solutions
In the genetic association studies described above, a variety of alleles have been associated with HAND; however, few findings have been reliably replicated. One possible reason for the lack of replicability of findings is the use of different neurobehavioral phenotypes across studies. Many used the diagnosis of HIV-associated dementia (HAD) as the phenotype 21,24,29,41,208, which is a largely unreliable behavioral phenotype8. Others have used composite measures of global neurocognitive functioning, usually derived from a comprehensive battery of neuropsychological tests 22,32,34. The major drawback of these phenotypes, particularly HAD, is that they are complex, and influenced by a number of environmental, psychometric, and endogenous factors. The numerous non-genetic contributors to variance in these measures (e.g., measurement error) makes them less suitable targets for genetic analysis, especially when effect sizes for genetic associations are generally small. Further, global measures of neurocognitive functioning run the risk of missing domain-specific associations with genes of interest. Therefore, domain specific composite scores (e.g., memory or processing speed) may be more suitable cognitive phenotypes for use in genetic analysis. Domain specific composite scores also have greater test-retest reliability when compared to individual measures, making them more attractive for creating progression phenotypes. In addition, the use of neurocognitive measures, many of which have demonstrated heritability, offers more precision and a more easily replicable phenotype across studies as compared to diagnostic categories.
Unlike the genomic studies, the majority of transcriptome studies have utilized encephalitis (either SIVE or HIVE) as their disease phenotype. In the pre-cART era, HIVE was considered the neuropathological basis of HAND, which usually manifested in the more severe forms of HAD and MCMD. However, in the current era, this relationship is not valid. As described by Everall et al 209, among 589 brains from the NNTC cohort, HIV-related pathology was observed in only 17.5% (11% with classic HIVE, 5% with microglial nodular encephalitis or aseptic leptomenningitis, and 1.5% with HIV-leukoencephalopathy), despite the fact that 88% of the sample had been diagnosed with HAND at some point pre-mortem. Instead, for the vast majority of HAND cases in the current era of widespread cART use, which are mild-to-moderate in severity, the neuropathogenesis of HAND is likely due to neuronal dysfunction caused by a mild and chronic neuroinflammatory state 161,166,209,210. This change may have been best demonstrated recently by Gelman et al., who compared transcription changes of those who had pre-mortem HAND without evidence of post-mortem HIVE to that of individuals with pre-mortem HAND who also showed post-mortem HIVE. The two groups had very distinctive transcriptome profiles despite a similar behavioral phenotype. This study underscores the need to utilize currently relevant disease phenotypes.
Due to these difficulties in utilizing neuropsychological tests, or diagnoses based primarily on such tests, as phenotypic measures of HAND, some have explored alternative outcome measures. Among these are various neuroimaging indices. While beyond the scope of this review, a variety of MRI and MRS markers have been associated with HIV disease progression, neurocognitive impairment, and response to putative treatments211-216. Still, and perhaps owing to the more widespread neuroimflammatory process underlying HAND, neuroimaging has not yet yielded a reliable biomarker that could be used as a phenotype in genetic and molecular biological studies, although this is likely to change. Another possible target is neuropathological changes quantified via immunohistochemistry. As such, new neuropathological markers have been sought to explain the behavioral manifestations characteristic of HAND. Among these is dendritic simplification 217, or a combination of synaptic and dendritic markers. For example, Moore et al found that a combined neuropathological phenotype consisting synaptodendritic neurodegeneration, as measured by synaptophysin (SYN) and microtubule-associated protein—2 (MAP2), is associated with HAND across both subcortical and cortical brain regions 9. Others have reported markers of β-amyloid deposition in the frontal cortex in HIV+ individuals 218-221. Whether or not this protein aggregation is etiologically related to MAP2 and SYN remains unclear. In addition to these markers of neurodegeneration and abnormal protein aggregation, markers of neuroinflammation have also been shown to be associated with neurocognitive impairment or HIV-related brain dysfunction 58,222,223. Indeed, macrophage proliferation, microglial activation, astrocytes activation, and increased chemokine levels have all been found within the CSF and brain of HIV+ individuals 11,35,36,161,166,210,224-226. While these may all represent candidates for neuropathogenic processes underlying HAND, new methods are necessary to determine which ones are relevant. Towards that end, an innovative approach for simultaneously determining which neuropathological markers are HAND-relevant and which genetic susceptibility loci influence HAND is to examine the association between genotype, neuropathological change, and behavioral outcomes. In this scenario, neuropathological changes (or neurophysiological in the case of neuroimaging) are considered intermediate phenotypes. Put more simply, if HAND is considered at its most basic level to be the end result of a sequence of physiological events that commences with HIV-induced cellular changes that are modified by genetic factors, then determining the extent to which known genetic susceptibility loci for HAND perturb candidate neuropathological and neurophysiological intermediate phenotypes may reveal which ones are most relevant. In this context, neuropathological intermediate phenotypes are less prone to exogenous factors and have a stronger association with genetic susceptibility loci than the neurobehavioral phenotypes (e.g., cognitive and functional deficits). As an example, the genetic susceptibility loci found to be associated with HAND, and discussed above, modify host immune response and also modify risk for HAND. It logically follows that these genetic variants must also modify the pathophysiological pathways linking immune response and HAND. This line of investigation is important for the elucidation of HAND neuropathogenesis, yet remains almost completely unexplored. This is not surprising, as this approach has only recently been successfully utilized in genetic association studies of Alzheimer’s disease 227,228. For example, the association between ApoE genotype and Alzheimer’s-related cognitive impairment was shown to be mediated by a sequential cascade of amyloid plaque formation and subsequent development of neurofibrillary tangle pathology 227-230. Relevant to HAND, to our knowledge only three studies has examined the relationship between genetic susceptibility loci and neuropathological outcomes. Sato-Matsumura et al. 231, with a sample of 44 AIDS patients with autopsy-verified HIVE or HIV-leukoencephalopathy and 30 AIDS patients without these neuropathologies, did not find an association between TNF-alpha genotype at rs1800629 and either of the neuropathological conditions. Diaz-Arrastia et al 78 assessed for HIVE or vacuolar myelopathy in the brains of 270 HIV+ individuals who died with AIDS between 1989 and 1996. Neurocognitive functioning and HAND were not considered. They determined presence of microglial nodules, multinucleated giant cells, myelin pallor, and vacuolar myelopathy in the brains and/or spinal cords. None of the alleles examined were associated with the presence of these markers. More recently, Soontornniyomkij et al. found that ApoE ε4 and older age were independently associated with increased the likelihood of cerebral amyloid β-plaque deposition in HIV+ adults66. While the amyloid β-plaques in HIV brains were immunohistologically different from those in brains of individuals who died with symptomatic Alzheimer’s disease, this study is the first to provide a link between genotype and neuropathological findings in HIV. However, neither study considered clinical manifestation of HAND in their design. A preliminary analysis of neuropathological, neuropsychological, and genetic data from a combined dataset from Levine et al 42 and Moore et al 9, indicates that the MCP-1 marker (rs1024611) is associated with synaptodendritic simplification, and also that synaptodendritic simplification is associated with neurocognitive functioning, suggesting that a neuropathogenic link between genotype, neuropathology, and clinical HAND may be possible232. This line of inquiry is currently being pursued with funding from the National Institute of Mental Health (R01MH096648-Levine & Moore).
5.2. Systems biologic analysis of ‘omics data – Weighted co-expression network analysis
HAND represents perhaps the most complex neurocognitive syndrome due to it heterogeneous pathogenesis, frequent co-morbidities, and lack of reliable biomarkers. While many genomic data sets have been generated (e.g., SNP marker data, gene expression data), a coherent pattern that might illuminate central pathways for pharmaceutical targeting has not yet materialized. Like the story of the blind men and the elephant, we have available to us diverse perspectives and data, but these have not been pieced together effectively. Looking forward, integrating these complementary data in a biologically meaningful fashion poses a challenge. For this purpose, systems biologic or network based approaches offer great promise.
Networks constructed from high-throughput data often reflect aspects of the underlying biology. For example, it has been observed that co-expressed genes tend to be also functionally related, and that clusters of co-expressed genes are often strongly enriched in specific functional categories or cell type markers 147. Transcriptional studies of various diseases have often found clusters of genes, termed modules, which correlate with disease status. For example, our WGCNA approach is a widely used systems biologic approach for finding disease related co-expression modules and intramodular hub genes 146,175,233. WGCNA can be interpreted as a step-wise data reduction technique, which (a) starts from the level of tens of thousands of variables (e.g., gene expression probes), (b) identifies biologically interesting modules (e.g. based on their association with disease status), (c) represents the modules by their centroids (e.g., eigenvectors or intramodular hubs), (d) uses intramodular connectivity (also known as module membership measure or kME) to annotate genes with respect to module membership, and (e) combines gene significance and module membership measures for identifying significant hub nodes. The module centric analysis alleviates the multiple testing problem inherent in high dimensional data. Instead of relating thousands of variables to disease status, it only correlates a few dozen modules to disease status. Highly connected intramodular hub genes can be interpreted as module centroids that best represent the module234. Often highly connected hub nodes in a network have been found to be important for the network’s functioning235.
Disease related co-expression modules may correspond to pathways or cell types131,175,236. Co-expression networks formed on the basis of pairwise correlation coefficients between genes are a special case of a correlation networks. Weighted correlation network analysis has not only been used for analyzing gene expression data but also for a variety of other "omics" data (e.g. microRNA data, DNA methylation data, and proteomics data)237,238. We recently compared several approaches for measuring co-expression relationships and found that a robust measure of the correlation (referred to as biweight midcorrelation) often outperforms other measures (e.g. based on mutual information) probably because it avoids the pitfalls of over-fitting239.
When selecting biomarkers (e.g., gene expression profile) for a clinical trait (e.g. disease status or survival time), the traditional (marginal) method is to base the selection on the statistical association of the individual candidate biomarkers with the disease (e.g., finding differentially expressed genes using a Student t-test or fold change criterion). Network approaches have emerged as an increasingly used alternative to these traditional analysis methods (as the previous sections illustrate in the context of HAND), but their use for selecting biologically or clinically important genes has not been explored extensively, in particular in the context of aggregating evidence from multiple data sets (meta-analysis). Many biostatisticians wonder when to use (intramodular) network connectivity instead of a more traditional statistical significance measure (e.g., a marginal meta-analysis method). To address this question, we recently compared hub gene selection (based on module membership to consensus modules) to marginal meta-analysis which considered each gene in isolation. Our comprehensive empirical studies revealed that the data analysis strategy (networks versus marginal approach) should be informed by the research goal240. When it comes to gaining biological insights regarding molecular processes then a co-expression-based meta-analysis method (consensus module analysis) typically perform much better than traditional marginal approaches. But when it comes to validation success of statistical associations with the clinical outcome, then traditional marginal meta-analysis methods perform as good as (if not better) than co-expression based approaches. Thus, when it comes to identifying highly significant and reproducible biomarkers, standard marginal approaches will work well. However, while standard approaches are good at detecting the low hanging fruits they often fail to identify weak and subtle effects. Consensus network based methods that focus on intramodular hubs can successfully tease out weak signals. Due to the heterogeneous and multifaceted nature of HAND pathogenesis, and the difficulties encountered thus far in validating robust biomarkers, such network based methods may be most fruitful.
6. Conclusion
Genetic, transcriptomic, and epigenetic studies have yielded a wealth of information about the neuropathogenesis of HAND. Intersecting results across these three methodologies indicate, for example, mitochondrial dysfunction, which has also been implicated in peripheral neuropathy, another common NeuroAIDS condition, perhaps opening a novel avenue of research for HAND therapeutics241-243. HAND is a behavioral syndrome for which the criteria will continue to evolve. As such, the utility of HAND as a phenotype for genetic and molecular biology studies will continue to yield inconsistent results. Emerging intermediate phenotype candidates, for which more robust associations with genetic and molecular markers might be found, include neuropathological (e.g., immunohistochemical markers) and neuroimaging markers. This, in combination with systems biological methods for data reduction and interpretation, represent an exciting and fruitful direction for investigating HAND neuropathogenesis. As more researchers adopt this approach, significant progress in delineating the biological pathways underlying HIV-related neurocognitive impairment is likely to be seen, and as a result will further improve diagnostic nosology for HAND.
Acknowledgements
Thank you to our research participants and colleagues from the Multicenter AIDS Cohort Study and National NeuroAIDS Tissue Consortium. A special thanks to Natasha Nemanim for her editorial assistance.
Source of Funding Research results presented in this review were made possible by the following grants: Clinical and Translational Science Institute & Center for AIDS Research at UCLA UL1TR000124 (Horvath); National Institute for Drug Abuse R01DA030913 (Levine and Horvath); UCLA AIDS Institute & Center for AIDS Research AI28697 (Freimer & Levine); National Institute for Drug Abuse R03DA026099 (Levine); California HIV/AIDS Research Program ID06-LA-187 (Levine).
Footnotes
Conflicts of Interest No conflicts of interest to report.
Contributor Information
Andrew J. Levine, Department of Neurology National Neurological AIDS Bank David Geffen School of Medicine at the University of California, Los Angeles.
Stella E. Panos, Department of Psychiatry and Biobehavioral Science David Geffen School of Medicine at the University of California, Los Angeles SPanos@mednet.ucla.edu
Steve Horvath, Department of Human Genetics David Geffen School of Medicine at the University of California, Los Angeles Department of Biostatistics, University of California, Los Angeles shorvath@mednet.ucla.edu
References
- 1.Navia BA, Jordan BD, Price RW. The AIDS dementia complex: I. Clinical features. Ann Neurol. 1986 Jun;19(6):517–524. doi: 10.1002/ana.410190602. [DOI] [PubMed] [Google Scholar]
- 2.Bornstein RA, Nasrallah HA, Para MF, Whitacre CC, Rosenberger P, Fass RJ. Neuropsychological performance in symptomatic and asymptomatic HIV infection. Aids. 1993 Apr;7(4):519–524. doi: 10.1097/00002030-199304000-00011. [DOI] [PubMed] [Google Scholar]
- 3.Antinori A, Arendt G, Becker JT, et al. Updated research nosology for HIV-associated neurocognitive disorders. Neurology. 2007 Oct 30;69(18):1789–1799. doi: 10.1212/01.WNL.0000287431.88658.8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gisslen M, Price RW, Nilsson S. The definition of HIV-associated neurocognitive disorders: are we overestimating the real prevalence? BMC Infect Dis. 2011;11:356. doi: 10.1186/1471-2334-11-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schretlen DJ, Munro CA, Anthony JC, Pearlson GD. Examining the range of normal intraindividual variability in neuropsychological test performance. J Int Neuropsychol Soc. 2003 Sep;9(6):864–870. doi: 10.1017/S1355617703960061. [DOI] [PubMed] [Google Scholar]
- 6.Blackstone K, Moore DJ, Franklin DR, et al. Defining neurocognitive impairment in HIV: deficit scores versus clinical ratings. Clin Neuropsychol. 2012;26(6):894–908. doi: 10.1080/13854046.2012.694479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgello S, Gelman BB, Kozlowski PB, et al. The National NeuroAIDS Tissue Consortium: a new paradigm in brain banking with an emphasis on infectious disease. Neuropathol Appl Neurobiol. 2001 Aug;27(4):326–335. doi: 10.1046/j.0305-1846.2001.00334.x. [DOI] [PubMed] [Google Scholar]
- 8.Woods SP, Rippeth JD, Frol AB, et al. Interrater reliability of clinical ratings and neurocognitive diagnoses in HIV. J Clin Exp Neuropsychol. 2004 Sep;26(6):759–778. doi: 10.1080/13803390490509565. [DOI] [PubMed] [Google Scholar]
- 9.Moore DJ, Masliah E, Rippeth JD, et al. Cortical and subcortical neurodegeneration is associated with HIV neurocognitive impairment. Aids. 2006 Apr 4;20(6):879–887. doi: 10.1097/01.aids.0000218552.69834.00. [DOI] [PubMed] [Google Scholar]
- 10.Everall I, Vaida F, Khanlou N, et al. Cliniconeuropathologic correlates of human immunodeficiency virus in the era of antiretroviral therapy. J Neurovirol. 2009 Sep;15(5-6):360–370. doi: 10.3109/13550280903131915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letendre SL, Zheng JC, Kaul M, et al. Chemokines in cerebrospinal fluid correlate with cerebral metabolite patterns in HIV-infected individuals. J Neurovirol. 2011 Feb;17(1):63–69. doi: 10.1007/s13365-010-0013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alkhatib G, Combadiere C, Broder CC, et al. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996 Jun 28;272(5270):1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 13.Alkhatib G, Broder CC, Berger EA. Cell type-specific fusion cofactors determine human immunodeficiency virus type 1 tropism for T-cell lines versus primary macrophages. J Virol. 1996 Aug;70(8):5487–5494. doi: 10.1128/jvi.70.8.5487-5494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane BR, King SR, Bock PJ, Strieter RM, Coffey MJ, Markovitz DM. The C-X-C chemokine IP-10 stimulates HIV-1 replication. Virology. 2003 Mar 1;307(1):122–134. doi: 10.1016/s0042-6822(02)00045-4. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez E, Dhanda R, Bamshad M, et al. Global survey of genetic variation in CCR5, RANTES, and MIP-1alpha: impact on the epidemiology of the HIV-1 pandemic. Proc Natl Acad Sci U S A. 2001 Apr 24;98(9):5199–5204. doi: 10.1073/pnas.091056898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez E, Kulkarni H, Bolivar H, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005 Mar 4;307(5714):1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 17.Kaul M, Lipton SA. Experimental and potential future therapeutic approaches for HIV-1 associated dementia targeting receptors for chemokines, glutamate and erythropoietin. Neurotox Res. 2005 Oct;8(1-2):167–186. doi: 10.1007/BF03033828. [DOI] [PubMed] [Google Scholar]
- 18.Weiss JM, Nath A, Major EO, Berman JW. HIV-1 Tat induces monocyte chemoattractant protein-1-mediated monocyte transmigration across a model of the human blood-brain barrier and up-regulates CCR5 expression on human monocytes. J Immunol. 1999 Sep 1;163(5):2953–2959. [PubMed] [Google Scholar]
- 19.Zheng J, Thylin MR, Cotter RL, et al. HIV-1 infected and immune competent mononuclear phagocytes induce quantitative alterations in neuronal dendritic arbor: relevance for HIV-1-associated dementia. Neurotox Res. 2001 Oct;3(5):443–459. doi: 10.1007/BF03033203. [DOI] [PubMed] [Google Scholar]
- 20.Zheng J, Thylin MR, Persidsky Y, et al. HIV-1 infected immune competent mononuclear phagocytes influence the pathways to neuronal demise. Neurotox Res. 2001 Oct;3(5):461–484. doi: 10.1007/BF03033204. [DOI] [PubMed] [Google Scholar]
- 21.Pemberton LA, Stone E, Price P, van Bockxmeer F, Brew BJ. The relationship between ApoE, TNFA, IL1a, IL1b and IL12b genes and HIV-1-associated dementia. HIV Med. 2008 Oct;9(8):677–680. doi: 10.1111/j.1468-1293.2008.00614.x. [DOI] [PubMed] [Google Scholar]
- 22.Spector SA, Singh KK, Gupta S, et al. APOE epsilon4 and MBL-2 O/O genotypes are associated with neurocognitive impairment in HIV-infected plasma donors. AIDS. Jun 19;24(10):1471–1479. doi: 10.1097/QAD.0b013e328339e25c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine AJ, Singer EJ, Shapshak P. The Role of Host Genetics in the Susceptibility for HIV-associated Neurocognitive Disorders. AIDS Behav. 2009 Feb;13(1):118–132. doi: 10.1007/s10461-008-9360-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine AJ, Singer EJ, Sinsheimer J.S, Hinkin, C.H., Papp J, Dandekar S, Giovanelli A, Shapshak P. CCL3 genotype and current depression increase risk of HIV-associated dementia. Neurobehavioral HIV Medicine. 2009 Nov 16;1:1–7. doi: 10.2147/nbhiv.s6820. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mummidi S, Okulicz JF, Wright EJ, He W, Ahuja SK. Genetic susceptibilities for NeuroAIDS. In: Gendelman HE, Grant I, Everall IP, Fox HS, Gelbard HA, Lipton SA, Swindells S, editors. The Neurology of AIDS. 3rd ed Oxford University Press; New York: 2012. pp. 71–89. [Google Scholar]
- 26.Kaul M, Ma Q, Medders KE, Desai MK, Lipton SA. HIV-1 coreceptors CCR5 and CXCR4 both mediate neuronal cell death but CCR5 paradoxically can also contribute to protection. Cell Death Differ. 2007 Feb;14(2):296–305. doi: 10.1038/sj.cdd.4402006. [DOI] [PubMed] [Google Scholar]
- 27.Liu H, Chao D, Nakayama EE, et al. Polymorphism in RANTES chemokine promoter affects HIV-1 disease progression. Proc Natl Acad Sci U S A. 1999 Apr 13;96(8):4581–4585. doi: 10.1073/pnas.96.8.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samson M, Libert F, Doranz BJ, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996 Aug 22;382(6593):722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 29.Boven LA, van der Bruggen T, van Asbeck BS, Marx JJ, Nottet HS. Potential role of CCR5 polymorphism in the development of AIDS dementia complex. FEMS Immunol Med Microbiol. 1999 Dec;26(3-4):243–247. doi: 10.1111/j.1574-695X.1999.tb01395.x. [DOI] [PubMed] [Google Scholar]
- 30.van Rij RP, Portegies P, Hallaby T, et al. Reduced prevalence of the CCR5 delta32 heterozygous genotype in human immunodeficiency virus-infected individuals with AIDS dementia complex. J Infect Dis. 1999 Sep;180(3):854–857. doi: 10.1086/314940. [DOI] [PubMed] [Google Scholar]
- 31.Spector SA, Singh KK, Gupta S, et al. APOE epsilon4 and MBL-2 O/O genotypes are associated with neurocognitive impairment in HIV-infected plasma donors. AIDS. 2010 Jun 19;24(10):1471–1479. doi: 10.1097/QAD.0b013e328339e25c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh KK, Ellis RJ, Marquie-Beck J, et al. CCR2 polymorphisms affect neuropsychological impairment in HIV-1-infected adults. J Neuroimmunol. 2004 Dec;157(1-2):185–192. doi: 10.1016/j.jneuroim.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 33.Bol SM, Booiman T, van Manen D, et al. Single nucleotide polymorphism in gene encoding transcription factor Prep1 is associated with HIV-1-associated dementia. PLoS One. 2012;7(2):e30990. doi: 10.1371/journal.pone.0030990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh KK, Barroga CF, Hughes MD, et al. Genetic influence of CCR5, CCR2, and SDF1 variants on human immunodeficiency virus 1 (HIV-1)-related disease progression and neurological impairment, in children with symptomatic HIV-1 infection. J Infect Dis. 2003 Nov 15;188(10):1461–1472. doi: 10.1086/379038. [DOI] [PubMed] [Google Scholar]
- 35.Eugenin EA, Osiecki K, Lopez L, Goldstein H, Calderon TM, Berman JW. CCL2/monocyte chemoattractant protein-1 mediates enhanced transmigration of human immunodeficiency virus (HIV)-infected leukocytes across the blood-brain barrier: a potential mechanism of HIV-CNS invasion and NeuroAIDS. J Neurosci. 2006 Jan 25;26(4):1098–1106. doi: 10.1523/JNEUROSCI.3863-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conant K, Garzino-Demo A, Nath A, et al. Induction of monocyte chemoattractant protein-1 in HIV-1 Tat-stimulated astrocytes and elevation in AIDS dementia. Proceedings of the National Academy of Sciences of the United States of America. 1998 Mar 17;95(6):3117–3121. doi: 10.1073/pnas.95.6.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelder W, McArthur JC, Nance-Sproson T, McClernon D, Griffin DE. Beta-chemokines MCP-1 and RANTES are selectively increased in cerebrospinal fluid of patients with human immunodeficiency virus-associated dementia. Ann Neurol. 1998 Nov;44(5):831–835. doi: 10.1002/ana.410440521. [DOI] [PubMed] [Google Scholar]
- 38.Lehmann MH, Masanetz S, Kramer S, Erfle V. HIV-1 Nef upregulates CCL2/MCP-1 expression in astrocytes in a myristoylation- and calmodulin-dependent manner. J Cell Sci. 2006 Nov 1;119(Pt 21):4520–4530. doi: 10.1242/jcs.03231. [DOI] [PubMed] [Google Scholar]
- 39.McDermott DH, Yang Q, Kathiresan S, et al. CCL2 polymorphisms are associated with serum monocyte chemoattractant protein-1 levels and myocardial infarction in the Framingham Heart Study. Circulation. 2005 Aug 23;112(8):1113–1120. doi: 10.1161/CIRCULATIONAHA.105.543579. [DOI] [PubMed] [Google Scholar]
- 40.Letendre S, Marquie-Beck J, Singh KK, et al. The monocyte chemotactic protein-1 -2578G allele is associated with elevated MCP-1 concentrations in cerebrospinal fluid. J Neuroimmunol. 2004 Dec;157(1-2):193–196. doi: 10.1016/j.jneuroim.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 41.Gonzalez E, Rovin BH, Sen L, et al. HIV-1 infection and AIDS dementia are influenced by a mutant MCP-1 allele linked to increased monocyte infiltration of tissues and MCP-1 levels. Proceedings of the National Academy of Sciences of the United States of America. 2002 Oct 15;99(21):13795–13800. doi: 10.1073/pnas.202357499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine AJ, Singer EJ, Sinsheimer JS, et al. CCL3 genotype and current depression increase risk of HIV-associated dementia. Neurobehav HIV Med. 2009 Nov;1:1–7. doi: 10.2147/nbhiv.s6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith MW, Dean M, Carrington M, et al. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997 Aug 15;277(5328):959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 44.Schmidtmayerova H, Nottet HS, Nuovo G, et al. Human immunodeficiency virus type 1 infection alters chemokine beta peptide expression in human monocytes: implications for recruitment of leukocytes into brain and lymph nodes. Proceedings of the National Academy of Sciences of the United States of America. 1996 Jan 23;93(2):700–704. doi: 10.1073/pnas.93.2.700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Modi WS, Lautenberger J, An P, et al. Genetic variation in the CCL18-CCL3-CCL4 chemokine gene cluster influences HIV Type 1 transmission and AIDS disease progression. American journal of human genetics. 2006 Jul;79(1):120–128. doi: 10.1086/505331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wesselingh SL, Takahashi K, Glass JD, McArthur JC, Griffin JW, Griffin DE. Cellular localization of tumor necrosis factor mRNA in neurological tissue from HIV-infected patients by combined reverse transcriptase/polymerase chain reaction in situ hybridization and immunohistochemistry. J Neuroimmunol. 1997 Apr;74(1-2):1–8. doi: 10.1016/s0165-5728(96)00160-9. [DOI] [PubMed] [Google Scholar]
- 47.Yeh MW, Kaul M, Zheng J, et al. Cytokine-stimulated, but not HIV-infected, human monocyte-derived macrophages produce neurotoxic levels of l -cysteine. J Immunol. 2000 Apr 15;164(8):4265–4270. doi: 10.4049/jimmunol.164.8.4265. [DOI] [PubMed] [Google Scholar]
- 48.Wesselingh SL, Power C, Glass JD, et al. Intracerebral cytokine messenger RNA expression in acquired immunodeficiency syndrome dementia. Ann Neurol. 1993 Jun;33(6):576–582. doi: 10.1002/ana.410330604. [DOI] [PubMed] [Google Scholar]
- 49.Chao CC, Hu S. Tumor necrosis factor-alpha potentiates glutamate neurotoxicity in human fetal brain cell cultures. Dev Neurosci. 1994;16(3-4):172–179. doi: 10.1159/000112104. [DOI] [PubMed] [Google Scholar]
- 50.Benos DJ, McPherson S, Hahn BH, Chaikin MA, Benveniste EN. Cytokines and HIV envelope glycoprotein gp120 stimulate Na+/H+ exchange in astrocytes. J Biol Chem. 1994 May 13;269(19):13811–13816. [PubMed] [Google Scholar]
- 51.Selmaj K, Raine CS. Tumor necrosis factor mediates myelin damage in organotypic cultures of nervous tissue. Ann N Y Acad Sci. 1988;540:568–570. doi: 10.1111/j.1749-6632.1988.tb27175.x. [DOI] [PubMed] [Google Scholar]
- 52.Gelbard HA, Nottet HS, Swindells S, et al. Platelet-activating factor: a candidate human immunodeficiency virus type 1-induced neurotoxin. J Virol. 1994 Jul;68(7):4628–4635. doi: 10.1128/jvi.68.7.4628-4635.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brabers NA, Nottet HS. Role of the pro-inflammatory cytokines TNF-alpha and IL-1beta in HIV-associated dementia. Eur J Clin Invest. 2006 Jul;36(7):447–458. doi: 10.1111/j.1365-2362.2006.01657.x. [DOI] [PubMed] [Google Scholar]
- 54.Quasney MW, Zhang Q, Sargent S, Mynatt M, Glass J, McArthur J. Increased frequency of the tumor necrosis factor-alpha-308 A allele in adults with human immunodeficiency virus dementia. Ann Neurol. 2001 Aug;50(2):157–162. [PubMed] [Google Scholar]
- 55.Agace WW, Amara A, Roberts AI, et al. Constitutive expression of stromal derived factor-1 by mucosal epithelia and its role in HIV transmission and propagation. Curr Biol. 2000 Mar 23;10(6):325–328. doi: 10.1016/s0960-9822(00)00380-8. [DOI] [PubMed] [Google Scholar]
- 56.Asensio VC, Campbell IL. Chemokines in the CNS: plurifunctional mediators in diverse states. Trends Neurosci. 1999 Nov;22(11):504–512. doi: 10.1016/s0166-2236(99)01453-8. [DOI] [PubMed] [Google Scholar]
- 57.Zheng J, Thylin MR, Ghorpade A, et al. Intracellular CXCR4 signaling, neuronal apoptosis and neuropathogenic mechanisms of HIV-1-associated dementia. Journal of neuroimmunology. 1999 Aug 3;98(2):185–200. doi: 10.1016/s0165-5728(99)00049-1. [DOI] [PubMed] [Google Scholar]
- 58.Kaul M, Lipton SA. Chemokines and activated macrophages in HIV gp120-induced neuronal apoptosis. Proc Natl Acad Sci U S A. 1999 Jul 6;96(14):8212–8216. doi: 10.1073/pnas.96.14.8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaul M, Zheng J, Okamoto S, Gendelman HE, Lipton SA. HIV-1 infection and AIDS: consequences for the central nervous system. Cell Death Differ. 2005 Aug;12(Suppl 1):878–892. doi: 10.1038/sj.cdd.4401623. [DOI] [PubMed] [Google Scholar]
- 60.Winkler C, Modi W, Smith MW, et al. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC) Science. 1998 Jan 16;279(5349):389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 61.Chang L, Andres M, Sadino J, et al. Impact of apolipoprotein E epsilon4 and HIV on cognition and brain atrophy: Antagonistic pleiotropy and premature brain aging. NeuroImage. 2011;58(4):1017–1027. doi: 10.1016/j.neuroimage.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vitek MP, Brown CM, Colton CA. APOE genotype-specific differences in the innate immune response. Neurobiol Aging. 2009 Sep;30(9):1350–1360. doi: 10.1016/j.neurobiolaging.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andres MA, Feger U, Nath A, Munsaka S, Jiang CS, Chang L. APOE epsilon 4 allele and CSF APOE on cognition in HIV-infected subjects. Journal of neuroimmune pharmacology : the official journal of the Society on NeuroImmune Pharmacology. 2011;6(3):389–398. doi: 10.1007/s11481-010-9254-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Panos SE, Hinkin CH, Singer EJ, et al. Apolipoprotein-E genotype and human immunodeficiency virus-associated neurocognitive disorder: The modulating effects of older age and disease severity. Neurobehavioral HIV Medicine. 2013 doi: 10.2147/NBHIV.S39573. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Diaz-Arrastia R, Gong Y, Kelly C, Gelman B. Host genetic polymorphisms in human immunodeficiency virus?related neurologic disease. Journal of neurovirology. 2004;10(Suppl 1):67–73. doi: 10.1080/753312755. [DOI] [PubMed] [Google Scholar]
- 66.Soontornniyomkij V, Moore DJ, Gouaux B, et al. Cerebral beta-amyloid deposition predicts HIV-associated neurocognitive disorders in APOE epsilon4 carriers. AIDS. 2012 Nov 28;26(18):2327–2335. doi: 10.1097/QAD.0b013e32835a117c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corder EH, Robertson KR, Lannfelt L, et al. HIV-infected subjects with E4 allele for APOE have excess demential and peripheral neuropathy. Nature medicine. 1998;4(10):1182–1184. doi: 10.1038/2677. [DOI] [PubMed] [Google Scholar]
- 68.Joska JA, Combrinck M, Valcour VG, et al. Association between apolipoprotein E4 genotype and human immunodeficiency virus-associated dementia in younger adults starting antiretroviral therapy in South Africa. Journal of neurovirology. 2010;16(5):377–383. doi: 10.3109/13550284.2010.513365. [DOI] [PubMed] [Google Scholar]
- 69.Pemberton LA, Stone E, Price P, van Bockxmeer F, Brew BJ. The relationship between ApoE, TNFA, IL1a, IL1b and IL12b genes and HIV-1-associated dementia. HIV Med. 2008;9(8):677–680. doi: 10.1111/j.1468-1293.2008.00614.x. [DOI] [PubMed] [Google Scholar]
- 70.Pomara N, Belzer KD, Silva R, Cooper TB, Sidtis JJ. The apolipoprotein E epsilon4 allele and memory performance in HIV-1 seropositive subjects: differences at baseline but not after acute oral lorazepam challenge. Psychopharmacology. 2008;201(1):125–135. doi: 10.1007/s00213-008-1253-1. [DOI] [PubMed] [Google Scholar]
- 71.Spector SA, Singh KK, Gupta S, et al. APOE e4 and MBL-2 O/O genotypes are associated with neurocognitive impairment in HIV-infected plasma donors. AIDS. 2010;24(10):1471–1479. doi: 10.1097/QAD.0b013e328339e25c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun B, Abadjian L, Rempel H, Calosing C, Rothlind J, Pulliam L. Peripheral biomarkers do not correlate with cognitive impairment in highly active antiretroviral therapy-treated subjects with human immunodeficiency virus type 1 infection. Journal of neurovirology. 2010;16(2):115–124. doi: 10.3109/13550280903559789. [DOI] [PubMed] [Google Scholar]
- 73.Valcour VG, Shikuma C, Shiramizu B, et al. Age, apolipoprotein E4, and the risk of HIV dementia: the Hawaii Aging with HIV Cohort. Journal of neuroimmunology. 2004;157(1-2):197–202. doi: 10.1016/j.jneuroim.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 74.Burt TD, Agan BK, Marconi VC, et al. Apolipoprotein (apo) E4 enhances HIV-1 cell entry in vitro, and the APOE epsilon4/epsilon4 genotype accelerates HIV disease progression. Proc Natl Acad Sci U S A. 2008;105(25):8718–8723. doi: 10.1073/pnas.0803526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Corder EH, Galeazzi L, Franceschi C, et al. Differential course of HIV-1 infection and apolipoprotein E polymorphism. Central European Journal of Medicine. 2007;2(4):404–416. [Google Scholar]
- 76.Valcour VG, Shiramizu B, Shikuma C. Frequency of apolipoprotein E4 among older compared with younger HIV patients: support for detrimental effect of E4 on survival. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(41):E66. doi: 10.1073/pnas.0806919105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morgan EE, Woods SP, Letendre SL, et al. Apolipoprotein E4 genotype does not increase risk of HIV-associated neurocognitive disorders. J Neurovirol. 2013 Apr;19(2):150–156. doi: 10.1007/s13365-013-0152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Diaz-Arrastia R, Gong Y, Kelly CJ, Gelman BB. Host genetic polymorphisms in human immunodeficiency virus-related neurologic disease. J Neurovirol. 2004;10(Suppl 1):67–73. doi: 10.1080/753312755. [DOI] [PubMed] [Google Scholar]
- 79.Dunlop, Goplen AK, Liestol K, et al. HIV dementia and apolipoprotein E. ActaNeurol Scand. 1997;95:315–318. doi: 10.1111/j.1600-0404.1997.tb00217.x. [DOI] [PubMed] [Google Scholar]
- 80.Goplen AK, Liestol K, Dunlop O, Bruun JN, Maehlen J. Dementia in AIDS patients in Oslo; the role of HIV encephalitis and CMV encephalitis. Scand J Infect Dis. 2001;33(10):755–758. doi: 10.1080/003655401317074572. [DOI] [PubMed] [Google Scholar]
- 81.Burt TD, Agan BK, Marconi VC, et al. Apolipoprotein (apo) E4 enhances HIV-1 cell entry in vitro, and the APOE epsilon4/epsilon4 genotype accelerates HIV disease progression. Proc Natl Acad Sci U S A. 2008 Jun 24;105(25):8718–8723. doi: 10.1073/pnas.0803526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cutler RG, Haughey NJ, Tammara A, et al. Dysregulation of sphingolipid and sterol metabolism by ApoE4 in HIV dementia. Neurology. 2004;63:626–630. doi: 10.1212/01.wnl.0000134662.19883.06. [DOI] [PubMed] [Google Scholar]
- 83.Fraser IP, Koziel H, Ezekowitz RA. The serum mannose-binding protein and the macrophage mannose receptor are pattern recognition molecules that link innate and adaptive immunity. Semin Immunol. 1998 Oct;10(5):363–372. doi: 10.1006/smim.1998.0141. [DOI] [PubMed] [Google Scholar]
- 84.Garred P, Madsen HO, Balslev U, et al. Susceptibility to HIV infection and progression of AIDS in relation to variant alleles of mannose-binding lectin. Lancet. 1997 Jan 25;349(9047):236–240. doi: 10.1016/S0140-6736(96)08440-1. [DOI] [PubMed] [Google Scholar]
- 85.Singh KK, Lieser A, Ruan PK, Fenton T, Spector SA. An age-dependent association of mannose-binding lectin-2 genetic variants on HIV-1-related disease in children. J Allergy Clin Immunol. 2008 Jul;122(1):173–180. 180, e171–172. doi: 10.1016/j.jaci.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Singh KK, Nathamu S, Adame A, et al. Expression of mannose binding lectin in HIV-1-infected brain: implications for HIV-related neuronal damage and neuroAIDS. Neurobehav HIV Med. 2011 May 1;3:41–52. doi: 10.2147/NBHIV.S19969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lachman HM, Papolos DF, Saito T, Yu YM, Szumlanski CL, Weinshilboum RM. Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics. 1996 Jun;6(3):243–250. doi: 10.1097/00008571-199606000-00007. [DOI] [PubMed] [Google Scholar]
- 88.Bilder RM, Volavka J, Lachman HM, Grace AA. The catechol-O-methyltransferase polymorphism: relations to the tonic-phasic dopamine hypothesis and neuropsychiatric phenotypes. Neuropsychopharmacology. 2004 Nov;29(11):1943–1961. doi: 10.1038/sj.npp.1300542. [DOI] [PubMed] [Google Scholar]
- 89.Bilder RM, Volavka J, Czobor P, et al. Neurocognitive correlates of the COMT Val(158)Met polymorphism in chronic schizophrenia. Biol Psychiatry. 2002 Oct 1;52(7):701–707. doi: 10.1016/s0006-3223(02)01416-6. [DOI] [PubMed] [Google Scholar]
- 90.Egan MF, Goldberg TE, Kolachana BS, et al. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci U S A. 2001 Jun 5;98(12):6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nolan KA, Bilder RM, Lachman HM, Volavka J. Catechol O-methyltransferase Val158Met polymorphism in schizophrenia: differential effects of Val and Met alleles on cognitive stability and flexibility. Am J Psychiatry. 2004 Feb;161(2):359–361. doi: 10.1176/appi.ajp.161.2.359. [DOI] [PubMed] [Google Scholar]
- 92.Rosa A, Peralta V, Cuesta MJ, et al. New evidence of association between COMT gene and prefrontal neurocognitive function in healthy individuals from sibling pairs discordant for psychosis. Am J Psychiatry. 2004 Jun;161(6):1110–1112. doi: 10.1176/appi.ajp.161.6.1110. [DOI] [PubMed] [Google Scholar]
- 93.Malhotra AK, Kestler LJ, Mazzanti C, Bates JA, Goldberg T, Goldman D. A functional polymorphism in the COMT gene and performance on a test of prefrontal cognition. Am J Psychiatry. 2002 Apr;159(4):652–654. doi: 10.1176/appi.ajp.159.4.652. [DOI] [PubMed] [Google Scholar]
- 94.Goldberg TE, Egan MF, Gscheidle T, et al. Executive subprocesses in working memory: relationship to catechol-O-methyltransferase Val158Met genotype and schizophrenia. Arch Gen Psychiatry. 2003 Sep;60(9):889–896. doi: 10.1001/archpsyc.60.9.889. [DOI] [PubMed] [Google Scholar]
- 95.Mattay VS, Goldberg TE, Fera F, et al. Catechol O-methyltransferase val158-met genotype and individual variation in the brain response to amphetamine. Proc Natl Acad Sci U S A. 2003 May 13;100(10):6186–6191. doi: 10.1073/pnas.0931309100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hsieh PC, Yeh TL, Lee IH, et al. Correlation between errors on the Wisconsin Card Sorting Test and the availability of striatal dopamine transporters in healthy volunteers. J Psychiatry Neurosci. Mar;35(2):90–94. doi: 10.1503/jpn.090007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mozley LH, Gur RC, Mozley PD, Gur RE. Striatal dopamine transporters and cognitive functioning in healthy men and women. Am J Psychiatry. 2001 Sep;158(9):1492–1499. doi: 10.1176/appi.ajp.158.9.1492. [DOI] [PubMed] [Google Scholar]
- 98.Wang GJ, Chang L, Volkow ND, et al. Decreased brain dopaminergic transporters in HIV-associated dementia patients. Brain. 2004 Nov;127(Pt 11):2452–2458. doi: 10.1093/brain/awh269. [DOI] [PubMed] [Google Scholar]
- 99.Chang L, Wang GJ, Volkow ND, et al. Decreased brain dopamine transporters are related to cognitive deficits in HIV patients with or without cocaine abuse. Neuroimage. 2008 Aug 15;42(2):869–878. doi: 10.1016/j.neuroimage.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Prata DP, Mechelli A, Fu CH, et al. Epistasis between the DAT 3′ UTR VNTR and the COMT Val158Met SNP on cortical function in healthy subjects and patients with schizophrenia. Proc Natl Acad Sci U S A. 2009 Aug 11;106(32):13600–13605. doi: 10.1073/pnas.0903007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bertolino A, Blasi G, Latorre V, et al. Additive effects of genetic variation in dopamine regulating genes on working memory cortical activity in human brain. J Neurosci. 2006 Apr 12;26(15):3918–3922. doi: 10.1523/JNEUROSCI.4975-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.VanNess SH, Owens MJ, Kilts CD. The variable number of tandem repeats element in DAT1 regulates in vitro dopamine transporter density. BMC Genet. 2005;6:55. doi: 10.1186/1471-2156-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guillin O, Diaz J, Carroll P, Griffon N, Schwartz JC, Sokoloff P. BDNF controls dopamine D3 receptor expression and triggers behavioural sensitization. Nature. 2001 May 3;411(6833):86–89. doi: 10.1038/35075076. [DOI] [PubMed] [Google Scholar]
- 104.Mossner R, Daniel S, Albert D, et al. Serotonin transporter function is modulated by brain-derived neurotrophic factor (BDNF) but not nerve growth factor (NGF) Neurochem Int. 2000 Mar;36(3):197–202. doi: 10.1016/s0197-0186(99)00122-9. [DOI] [PubMed] [Google Scholar]
- 105.Nosheny RL, Mocchetti I, Bachis A. Brain-derived neurotrophic factor as a prototype neuroprotective factor against HIV-1-associated neuronal degeneration. Neurotox Res. 2005 Oct;8(1-2):187–198. doi: 10.1007/BF03033829. [DOI] [PubMed] [Google Scholar]
- 106.Nosheny RL, Ahmed F, Yakovlev A, et al. Brain-derived neurotrophic factor prevents the nigrostriatal degeneration induced by human immunodeficiency virus-1 glycoprotein 120 in vivo. Eur J Neurosci. 2007 Apr;25(8):2275–2284. doi: 10.1111/j.1460-9568.2007.05506.x. [DOI] [PubMed] [Google Scholar]
- 107.Egan MF, Kojima M, Callicott JH, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003 Jan 24;112(2):257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 108.Pezawas L, Verchinski BA, Mattay VS, et al. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004 Nov 10;24(45):10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mocchetti I, Nosheny RL, Tanda G, Ren K, Meyer EM. Brain-derived neurotrophic factor prevents human immunodeficiency virus type 1 protein gp120 neurotoxicity in the rat nigrostriatal system. Ann N Y Acad Sci. 2007 Dec;1122:144–154. doi: 10.1196/annals.1403.010. [DOI] [PubMed] [Google Scholar]
- 110.Kumar AM, Ownby RL, Waldrop-Valverde D, Fernandez B, Kumar M. Human immunodeficiency virus infection in the CNS and decreased dopamine availability: relationship with neuropsychological performance. J Neurovirol. 2011 Feb;17(1):26–40. doi: 10.1007/s13365-010-0003-4. [DOI] [PubMed] [Google Scholar]
- 111.Kumar AM, Fernandez J, Singer EJ, et al. Human immunodeficiency virus type 1 in the central nervous system leads to decreased dopamine in different regions of postmortem human brains. J Neurovirol. 2009 Jun 3;:1–18. doi: 10.1080/13550280902973952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gelman BB, Lisinicchia JG, Chen T, et al. Prefrontal dopaminergic and enkephalinergic synaptic accommodation in HIV-associated neurocognitive disorders and encephalitis. J Neuroimmune Pharmacol. 2012 Sep;7(3):686–700. doi: 10.1007/s11481-012-9345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gelman BB, Spencer JA, Holzer CE, 3rd, Soukup VM. Abnormal striatal dopaminergic synapses in National NeuroAIDS Tissue Consortium subjects with HIV encephalitis. J Neuroimmune Pharmacol. 2006 Dec;1(4):410–420. doi: 10.1007/s11481-006-9030-6. [DOI] [PubMed] [Google Scholar]
- 114.Levine AJ, Sinsheimer JS, Bilder R, Shapshak P, Singer EJ. Functional polymorphisms in dopamine-related genes: effect on neurocognitive functioning in HIV+ adults. J Clin Exp Neuropsychol. 2012;34(1):78–91. doi: 10.1080/13803395.2011.623118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bousman CA, Cherner M, Atkinson JH, et al. COMT Val158Met Polymorphism, Executive Dysfunction, and Sexual Risk Behavior in the Context of HIV Infection and Methamphetamine Dependence. Interdiscip Perspect Infect Dis. 2010:678648. doi: 10.1155/2010/678648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gupta S, Bousman CA, Chana G, et al. Dopamine receptor D3 genetic polymorphism (rs6280TC) is associated with rates of cognitive impairment in methamphetamine-dependent men with HIV: preliminary findings. J Neurovirol. 2011 Apr 14; doi: 10.1007/s13365-011-0028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Levine AJ, Reynolds S, Cox C, Miller E, Service S, Becker J, Sacktor N, Martin E, the Mutlicenter AIDS Cohort Study Longitudinal analysis of interactive effects of host genotype, stimulant use, and HIV status upon neurocognitive functioning. 11th Symposium of the International Society on NeuroVirology; New York, NY. May 29-June 2.2012. [Google Scholar]
- 118.Aouizerat BE, Pearce CL, Miaskowski C. The search for host genetic factors of HIV/AIDS pathogenesis in the post-genome era: progress to date and new avenues for discovery. Curr HIV/AIDS Rep. 2011 Mar;8(1):38–44. doi: 10.1007/s11904-010-0065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.An P, Winkler CA. Host genes associated with HIV/AIDS: advances in gene discovery. Trends Genet. 2010 Mar;26(3):119–131. doi: 10.1016/j.tig.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Belbin O, Carrasquillo MM, Crump M, et al. Investigation of 15 of the top candidate genes for late-onset Alzheimer’s disease. Hum. Genet. Mar. 2011;129(3):273–282. doi: 10.1007/s00439-010-0924-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Seshadri S, Fitzpatrick AL, Ikram MA, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010 May 12;303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Laumet G, Chouraki V, Grenier-Boley B, et al. Systematic analysis of candidate genes for Alzheimer’s disease in a French, genome-wide association study. J Alzheimers Dis. 2010;20(4):1181–1188. doi: 10.3233/JAD-2010-100126. [DOI] [PubMed] [Google Scholar]
- 123.Braskie MN, Jahanshad N, Stein JL, et al. Common Alzheimer’s disease risk variant within the CLU gene affects white matter microstructure in young adults. J Neurosci. 2011 May 4;31(18):6764–6770. doi: 10.1523/JNEUROSCI.5794-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ho AJ, Stein JL, Hua X, et al. A commonly carried allele of the obesity-related FTO gene is associated with reduced brain volume in the healthy elderly. Proc Natl Acad Sci U S A. 2010 May 4;107(18):8404–8409. doi: 10.1073/pnas.0910878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Levine AJ, Service S, Miller EN, et al. Genome-wide association study of neurocognitive impairment and dementia in HIV-infected adults. Am J Med Genet B Neuropsychiatr Genet. 2012 May 24; doi: 10.1002/ajmg.b.32071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Noorbakhsh F, Ramachandran R, Barsby N, et al. MicroRNA profiling reveals new aspects of HIV neurodegeneration: caspase-6 regulates astrocyte survival. FASEB J. 2010 Jun;24(6):1799–1812. doi: 10.1096/fj.09-147819. [DOI] [PubMed] [Google Scholar]
- 127.Gelman BB, Soukup VM, Schuenke KW, et al. Acquired neuronal channelopathies in HIV-associated dementia. J Neuroimmunol. 2004 Dec;157(1-2):111–119. doi: 10.1016/j.jneuroim.2004.08.044. [DOI] [PubMed] [Google Scholar]
- 128.Benos DJ, Hahn BH, Bubien JK, et al. Envelope glycoprotein gp120 of human immunodeficiency virus type 1 alters ion transport in astrocytes: implications for AIDS dementia complex. Proc Natl Acad Sci U S A. 1994 Jan 18;91(2):494–498. doi: 10.1073/pnas.91.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cantor RM, Lange K, Sinsheimer JS. Prioritizing GWAS results: A review of statistical methods and recommendations for their application. Am J Hum Genet. 2010 Jan;86(1):6–22. doi: 10.1016/j.ajhg.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shapshak P, Duncan R, Torres-Munoz JE, Duran EM, Minagar A, Petito CK. Analytic approaches to differential gene expression in AIDS versus control brains. Front Biosci. 2004 Sep 1;9:2935–2946. doi: 10.2741/1449. [DOI] [PubMed] [Google Scholar]
- 131.Presson AP, Sobel EM, Papp JC, et al. Integrated weighted gene co-expression network analysis with an application to chronic fatigue syndrome. BMC Syst Biol. 2008;2:95. doi: 10.1186/1752-0509-2-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Giri MS, Nebozhyn M, Showe L, Montaner LJ. Microarray data on gene modulation by HIV-1 in immune cells: 2000-2006. J Leukoc Biol. 2006 Nov;80(5):1031–1043. doi: 10.1189/jlb.0306157. [DOI] [PubMed] [Google Scholar]
- 133.Kim SY, Li J, Bentsman G, Brooks AI, Volsky DJ. Microarray analysis of changes in cellular gene expression induced by productive infection of primary human astrocytes: implications for HAD. J Neuroimmunol. 2004 Dec;157(1-2):17–26. doi: 10.1016/j.jneuroim.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 134.Kolson DL, Sabnekar P, Baybis M, Crino PB. Gene expression in TUNEL-positive neurons in human immunodeficiency virus-infected brain. J Neurovirol. 2004;10(Suppl 1):102–107. doi: 10.1080/753312760. [DOI] [PubMed] [Google Scholar]
- 135.Borjabad A, Brooks AI, Volsky DJ. Gene expression profiles of HIV-1-infected glia and brain: toward better understanding of the role of astrocytes in HIV-1-associated neurocognitive disorders. J Neuroimmune Pharmacol. Mar;5(1):44–62. doi: 10.1007/s11481-009-9167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Masliah E, Roberts ES, Langford D, et al. Patterns of gene dysregulation in the frontal cortex of patients with HIV encephalitis. J Neuroimmunol. 2004 Dec;157(1-2):163–175. doi: 10.1016/j.jneuroim.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 137.Salaria S, Badkoobehi H, Rockenstein E, et al. Toll-like receptor pathway gene expression is associated with human immunodeficiency virus-associated neurodegeneration. J Neurovirol. 2007 Dec;13(6):496–503. doi: 10.1080/13550280701558616. [DOI] [PubMed] [Google Scholar]
- 138.Everall I, Salaria S, Roberts E, et al. Methamphetamine stimulates interferon inducible genes in HIV infected brain. J Neuroimmunol. 2005 Dec 30;170(1-2):158–171. doi: 10.1016/j.jneuroim.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 139.Winkler JM, Chaudhuri AD, Fox HS. Translating the Brain Transcriptome in NeuroAIDS: From Non-human Primates to Humans. J Neuroimmune Pharmacol. 2012 Feb 28; doi: 10.1007/s11481-012-9344-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Roberts ES, Zandonatti MA, Watry DD, et al. Induction of pathogenic sets of genes in macrophages and neurons in NeuroAIDS. Am J Pathol. 2003 Jun;162(6):2041–2057. doi: 10.1016/S0002-9440(10)64336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gersten M, Alirezaei M, Marcondes MC, et al. An integrated systems analysis implicates EGR1 downregulation in simian immunodeficiency virus encephalitis-induced neural dysfunction. J. Neurosci. 2009 Oct 7;29(40):12467–12476. doi: 10.1523/JNEUROSCI.3180-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Borjabad A, Brooks AI, Volsky DJ. Gene expression profiles of HIV-1-infected glia and brain: toward better understanding of the role of astrocytes in HIV-1-associated neurocognitive disorders. J Neuroimmune Pharmacol. 2010 Mar;5(1):44–62. doi: 10.1007/s11481-009-9167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Winkler JM, Chaudhuri AD, Fox HS. Translating the brain transcriptome in neuroAIDS: from non-human primates to humans. J Neuroimmune Pharmacol. 2012 Jun;7(2):372–379. doi: 10.1007/s11481-012-9344-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gelman BB, Chen T, Lisinicchia JG, et al. The National NeuroAIDS Tissue Consortium Brain Gene Array: Two Types of HIV-Associated Neurocognitive Impairment. PLoS One. 2012;7(9):e46178. doi: 10.1371/journal.pone.0046178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Levine AJ, Miller JA, Shapshak P, et al. Systems analysis of human brain gene expression: mechanisms for HIV-associated neurocognitive impairment and common pathways with Alzheimer’s disease. BMC Med Genomics. 2013 Feb 13;6(1):4. doi: 10.1186/1755-8794-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Oldham MC, Konopka G, Iwamoto K, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008 Nov;11(11):1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Borjabad A, Volsky DJ. Common Transcriptional Signatures in Brain Tissue from Patients with HIV-Associated Neurocognitive Disorders, Alzheimer’s Disease, and Multiple Sclerosis. J Neuroimmune Pharmacol. 2012 Oct 12; doi: 10.1007/s11481-012-9409-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Borjabad A, Morgello S, Chao W, et al. Significant effects of antiretroviral therapy on global gene expression in brain tissues of patients with HIV-1-associated neurocognitive disorders. PLoS Pathog. 2011 Sep;7(9):e1002213. doi: 10.1371/journal.ppat.1002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Letendre S, Marquie-Beck J, Capparelli E, et al. Validation of the CNS Penetration-Effectiveness rank for quantifying antiretroviral penetration into the central nervous system. Arch Neurol. 2008 Jan;65(1):65–70. doi: 10.1001/archneurol.2007.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Marra CM, Zhao Y, Clifford DB, et al. Impact of combination antiretroviral therapy on cerebrospinal fluid HIV RNA and neurocognitive performance. AIDS. 2009 Jul 17;23(11):1359–1366. doi: 10.1097/QAD.0b013e32832c4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tozzi V, Balestra P, Bellagamba R, et al. Persistence of neuropsychologic deficits despite long-term highly active antiretroviral therapy in patients with HIV-related neurocognitive impairment: prevalence and risk factors. J Acquir Immune Defic Syndr. 2007 Jun 1;45(2):174–182. doi: 10.1097/QAI.0b013e318042e1ee. [DOI] [PubMed] [Google Scholar]
- 153.Cysique LA, Vaida F, Letendre S, et al. Dynamics of cognitive change in impaired HIV-positive patients initiating antiretroviral therapy. Neurology. 2009 Aug 4;73(5):342–348. doi: 10.1212/WNL.0b013e3181ab2b3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Marra CM, Lockhart D, Zunt JR, Perrin M, Coombs RW, Collier AC. Changes in CSF and plasma HIV-1 RNA and cognition after starting potent antiretroviral therapy. Neurology. 2003 Apr 22;60(8):1388–1390. doi: 10.1212/01.wnl.0000058768.73358.1a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Garvey L, Winston A, Walsh J, et al. Antiretroviral therapy CNS penetration and HIV-1-associated CNS disease. Neurology. 2011 Feb 22;76(8):693–700. doi: 10.1212/WNL.0b013e31820d8b0b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kahouadji Y, Dumurgier J, Sellier P, et al. Cognitive function after several years of antiretroviral therapy with stable central nervous system penetration score. HIV Med. 2012 Oct 4; doi: 10.1111/j.1468-1293.2012.01052.x. [DOI] [PubMed] [Google Scholar]
- 157.Pulliam L, Gascon R, Stubblebine M, McGuire D, McGrath MS. Unique monocyte subset in patients with AIDS dementia. Lancet. 1997 Mar 8;349(9053):692–695. doi: 10.1016/S0140-6736(96)10178-1. [DOI] [PubMed] [Google Scholar]
- 158.Ellery PJ, Tippett E, Chiu YL, et al. The CD16+ monocyte subset is more permissive to infection and preferentially harbors HIV-1 in vivo. J Immunol. 2007 May 15;178(10):6581–6589. doi: 10.4049/jimmunol.178.10.6581. [DOI] [PubMed] [Google Scholar]
- 159.Peluso R, Haase A, Stowring L, Edwards M, Ventura P. A Trojan Horse mechanism for the spread of visna virus in monocytes. Virology. 1985 Nov;147(1):231–236. doi: 10.1016/0042-6822(85)90246-6. [DOI] [PubMed] [Google Scholar]
- 160.Ancuta P, Moses A, Gabuzda D. Transendothelial migration of CD16+ monocytes in response to fractalkine under constitutive and inflammatory conditions. Immunobiology. 2004;209(1-2):11–20. doi: 10.1016/j.imbio.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 161.Kraft-Terry SD, Buch SJ, Fox HS, Gendelman HE. A coat of many colors: neuroimmune crosstalk in human immunodeficiency virus infection. Neuron. 2009 Oct 15;64(1):133–145. doi: 10.1016/j.neuron.2009.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Persidsky Y, Ghorpade A, Rasmussen J, et al. Microglial and astrocyte chemokines regulate monocyte migration through the blood-brain barrier in human immunodeficiency virus-1 encephalitis. Am J Pathol. 1999 Nov;155(5):1599–1611. doi: 10.1016/S0002-9440(10)65476-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Persidsky Y, Zheng J, Miller D, Gendelman HE. Mononuclear phagocytes mediate blood-brain barrier compromise and neuronal injury during HIV-1-associated dementia. J Leukoc Biol. 2000 Sep;68(3):413–422. [PubMed] [Google Scholar]
- 164.Cosenza MA, Zhao ML, Si Q, Lee SC. Human brain parenchymal microglia express CD14 and CD45 and are productively infected by HIV-1 in HIV-1 encephalitis. Brain Pathol. 2002 Oct;12(4):442–455. doi: 10.1111/j.1750-3639.2002.tb00461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kedzierska K, Crowe SM. The role of monocytes and macrophages in the pathogenesis of HIV-1 infection. Curr Med Chem. 2002 Nov;9(21):1893–1903. doi: 10.2174/0929867023368935. [DOI] [PubMed] [Google Scholar]
- 166.Glass JD, Fedor H, Wesselingh SL, McArthur JC. Immunocytochemical quantitation of human immunodeficiency virus in the brain: correlations with dementia. Ann Neurol. 1995 Nov;38(5):755–762. doi: 10.1002/ana.410380510. [DOI] [PubMed] [Google Scholar]
- 167.Adle-Biassette H, Chretien F, Wingertsmann L, et al. Neuronal apoptosis does not correlate with dementia in HIV infection but is related to microglial activation and axonal damage. Neuropathol Appl Neurobiol. 1999 Apr;25(2):123–133. doi: 10.1046/j.1365-2990.1999.00167.x. [DOI] [PubMed] [Google Scholar]
- 168.Lindl KA, Akay C, Wang Y, White MG, Jordan-Sciutto KL. Expression of the endoplasmic reticulum stress response marker, BiP, in the central nervous system of HIV-positive individuals. Neuropathol Appl Neurobiol. 2007 Dec;33(6):658–669. doi: 10.1111/j.1365-2990.2007.00866.x. [DOI] [PubMed] [Google Scholar]
- 169.Kaul M, Lipton SA. Mechanisms of neuronal injury and death in HIV-1 associated dementia. Curr HIV Res. 2006 Jul;4(3):307–318. doi: 10.2174/157016206777709384. [DOI] [PubMed] [Google Scholar]
- 170.Buckner CM, Calderon TM, Willams DW, Belbin TJ, Berman JW. Characterization of monocyte maturation/differentiation that facilitates their transmigration across the blood-brain barrier and infection by HIV: implications for NeuroAIDS. Cell Immunol. 2011;267(2):109–123. doi: 10.1016/j.cellimm.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pulliam L, Sun B, Rempel H. Invasive chronic inflammatory monocyte phenotype in subjects with high HIV-1 viral load. J Neuroimmunol. 2004 Dec;157(1-2):93–98. doi: 10.1016/j.jneuroim.2004.08.039. [DOI] [PubMed] [Google Scholar]
- 172.Sun B, Abadjian L, Rempel H, Calosing C, Rothlind J, Pulliam L. Peripheral biomarkers do not correlate with cognitive impairment in highly active antiretroviral therapy-treated subjects with human immunodeficiency virus type 1 infection. J Neurovirol. 2010 Mar;16(2):115–124. doi: 10.3109/13550280903559789. [DOI] [PubMed] [Google Scholar]
- 173.Pulliam L, Rempel H, Sun B, Abadjian L, Calosing C, Meyerhoff DJ. A peripheral monocyte interferon phenotype in HIV infection correlates with a decrease in magnetic resonance spectroscopy metabolite concentrations. AIDS. 2011 Sep 10;25(14):1721–1726. doi: 10.1097/QAD.0b013e328349f022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Levine AJ, Horvath S, Miller EN, et al. Transcriptome analysis of HIV-infected peripheral blood monocytes: Gene transcripts and networks associated with neurocognitive functioning. J Neuroimmunol. 2013 Sep 26; doi: 10.1016/j.jneuroim.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Horvath S, Zhang B, Carlson M, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A. 2006 Nov 14;103(46):17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yoshioka M, Shapshak P, Srivastava AK, et al. Expression of HIV-1 and interleukin-6 in lumbosacral dorsal root ganglia of patients with AIDS. Neurology. 1994 Jun;44(6):1120–1130. doi: 10.1212/wnl.44.6.1120. [DOI] [PubMed] [Google Scholar]
- 177.Pedersen KK, Pedersen M, Gaardbo JC, et al. Persisting Inflammation and Chronic Immune Activation but Intact Cognitive Function in HIV-infected patients After Long Term Treatment with Combination Antiretroviral Therapy. J Acquir Immune Defic Syndr. 2013 Feb 7; doi: 10.1097/QAI.0b013e318289bced. [DOI] [PubMed] [Google Scholar]
- 178.Perez DI, Gil C, Martinez A. Protein kinases CK1 and CK2 as new targets for neurodegenerative diseases. Med Res Rev. 2011 Nov;31(6):924–954. doi: 10.1002/med.20207. [DOI] [PubMed] [Google Scholar]
- 179.Janeczko K. The proliferative response of S-100 protein-positive glial cells to injury in the neonatal rat brain. Brain Res. 1991 Nov 8;564(1):86–90. doi: 10.1016/0006-8993(91)91355-5. [DOI] [PubMed] [Google Scholar]
- 180.Kuwabara K, Matsumoto M, Ikeda J, et al. Purification and characterization of a novel stress protein, the 150-kDa oxygen-regulated protein (ORP150), from cultured rat astrocytes and its expression in ischemic mouse brain. J Biol Chem. 1996 Mar 1;271(9):5025–5032. doi: 10.1074/jbc.271.9.5025. [DOI] [PubMed] [Google Scholar]
- 181.Ozawa K, Kondo T, Hori O, et al. Expression of the oxygen-regulated protein ORP150 accelerates wound healing by modulating intracellular VEGF transport. J Clin Invest. 2001 Jul;108(1):41–50. doi: 10.1172/JCI11772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tamatani M, Matsuyama T, Yamaguchi A, et al. ORP150 protects against hypoxia/ischemia-induced neuronal death. Nat Med. 2001 Mar;7(3):317–323. doi: 10.1038/85463. [DOI] [PubMed] [Google Scholar]
- 183.Zhao L, Rosales C, Seburn K, Ron D, Ackerman SL. Alteration of the unfolded protein response modifies neurodegeneration in a mouse model of Marinesco-Sjogren syndrome. Hum Mol Genet. 2010 Jan 1;19(1):25–35. doi: 10.1093/hmg/ddp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kounnas MZ, Moir RD, Rebeck GW, et al. LDL receptor-related protein, a multifunctional ApoE receptor, binds secreted beta-amyloid precursor protein and mediates its degradation. Cell. 1995 Jul 28;82(2):331–340. doi: 10.1016/0092-8674(95)90320-8. [DOI] [PubMed] [Google Scholar]
- 185.Liu Y, Jones M, Hingtgen CM, et al. Uptake of HIV-1 tat protein mediated by low-density lipoprotein receptor-related protein disrupts the neuronal metabolic balance of the receptor ligands. Nat Med. 2000 Dec;6(12):1380–1387. doi: 10.1038/82199. [DOI] [PubMed] [Google Scholar]
- 186.Kang DE, Saitoh T, Chen X, et al. Genetic association of the low-density lipoprotein receptor-related protein gene (LRP), an apolipoprotein E receptor, with late-onset Alzheimer’s disease. Neurology. 1997 Jul;49(1):56–61. doi: 10.1212/wnl.49.1.56. [DOI] [PubMed] [Google Scholar]
- 187.Bian L, Yang JD, Guo TW, et al. Association study of the A2M and LRP1 Genes with Alzheimer disease in the Han Chinese. Biol Psychiatry. 2005 Nov 1;58(9):731–737. doi: 10.1016/j.biopsych.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 188.Itoh K, Wakabayashi N, Katoh Y, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999 Jan 1;13(1):76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004 Nov;10(11):549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 190.Tanji K, Maruyama A, Odagiri S, et al. Keap1 is localized in neuronal and glial cytoplasmic inclusions in various neurodegenerative diseases. J Neuropathol Exp Neurol. 2013 Jan;72(1):18–28. doi: 10.1097/NEN.0b013e31827b5713. [DOI] [PubMed] [Google Scholar]
- 191.Zhao J, Redell JB, Moore AN, Dash PK. A novel strategy to activate cytoprotective genes in the injured brain. Biochem Biophys Res Commun. 2011 Apr 15;407(3):501–506. doi: 10.1016/j.bbrc.2011.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Reddy PV, Agudelo M, Atluri VS, Nair MP. Inhibition of nuclear factor erythroid 2-related factor 2 exacerbates HIV-1 gp120-induced oxidative and inflammatory response: role in HIV associated neurocognitive disorder. Neurochem Res. 2012 Aug;37(8):1697–1706. doi: 10.1007/s11064-012-0779-0. [DOI] [PubMed] [Google Scholar]
- 193.Lo SC, Li X, Henzl MT, Beamer LJ, Hannink M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006 Aug 9;25(15):3605–3617. doi: 10.1038/sj.emboj.7601243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Surh YJ, Kundu JK, Na HK. Nrf2 as a master redox switch in turning on the cellular signaling involved in the induction of cytoprotective genes by some chemopreventive phytochemicals. Planta medica. 2008 Oct;74(13):1526–1539. doi: 10.1055/s-0028-1088302. [DOI] [PubMed] [Google Scholar]
- 195.Letendre SL, Woods SP, Ellis RJ, et al. Lithium improves HIV-associated neurocognitive impairment. AIDS. 2006 Sep 11;20(14):1885–1888. doi: 10.1097/01.aids.0000244208.49123.1b. [DOI] [PubMed] [Google Scholar]
- 196.Guo L, Xing Y, Pan R, et al. Curcumin protects microglia and primary rat cortical neurons against HIV-1 gp120-mediated inflammation and apoptosis. PLoS One. 2013;8(8):e70565. doi: 10.1371/journal.pone.0070565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Roberts ES, Burudi EM, Flynn C, et al. Acute SIV infection of the brain leads to upregulation of IL6 and interferon-regulated genes: expression patterns throughout disease progression and impact on neuroAIDS. J Neuroimmunol. 2004 Dec;157(1-2):81–92. doi: 10.1016/j.jneuroim.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 198.Shapshak P, Duncan R, Minagar A, Rodriguez de la Vega P, Stewart RV, Goodkin K. Elevated expression of IFN-gamma in the HIV-1 infected brain. Front Biosci. 2004 May 1;9:1073–1081. doi: 10.2741/1271. [DOI] [PubMed] [Google Scholar]
- 199.Eletto D, Russo G, Passiatore G, et al. Inhibition of SNAP25 expression by HIV-1 Tat involves the activity of mir-128a. J Cell Physiol. 2008 Sep;216(3):764–770. doi: 10.1002/jcp.21452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Yelamanchili SV, Chaudhuri AD, Chen LN, Xiong H, Fox HS. MicroRNA-21 dysregulates the expression of MEF2C in neurons in monkey and human SIV/HIV neurological disease. Cell Death Dis. 2010;1:e77. doi: 10.1038/cddis.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tatro ET, Scott ER, Nguyen TB, et al. Evidence for Alteration of Gene Regulatory Networks through MicroRNAs of the HIV-infected brain: novel analysis of retrospective cases. PLoS One. 2010;5(4):e10337. doi: 10.1371/journal.pone.0010337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mukerjee R, Chang JR, Del Valle L, et al. Deregulation of microRNAs by HIV-1 Vpr protein leads to the development of neurocognitive disorders. J Biol Chem. 2011 Oct 7;286(40):34976–34985. doi: 10.1074/jbc.M111.241547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sun JM, Spencer VA, Chen HY, Li L, Davie JR. Measurement of histone acetyltransferase and histone deacetylase activities and kinetics of histone acetylation. Methods. 2003 Sep;31(1):12–23. doi: 10.1016/s1046-2023(03)00083-5. [DOI] [PubMed] [Google Scholar]
- 204.Thomas EA. Focal nature of neurological disorders necessitates isotype-selective histone deacetylase (HDAC) inhibitors. Mol Neurobiol. 2009 Aug;40(1):33–45. doi: 10.1007/s12035-009-8067-y. [DOI] [PubMed] [Google Scholar]
- 205.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov. 2008 Oct;7(10):854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 206.Saiyed ZM, Gandhi N, Agudelo M, et al. HIV-1 Tat upregulates expression of histone deacetylase-2 (HDAC2) in human neurons: implication for HIV-associated neurocognitive disorder (HAND) Neurochem Int. 2011 May;58(6):656–664. doi: 10.1016/j.neuint.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Perez-Santiago J PN, Letendre S, Ellis R, Gouaux B, Moore D, LeBlanc S, Rajagopal N, Zhang K, Woelk C. DNA methylation correlates with neurological decline in HIV-infected individuals. 19th Conference on Retroviruses and Opportunistic Infections; Seattle, Washington. 2012. [Google Scholar]
- 208.Valcour V, Shikuma C, Shiramizu B, et al. Age, apolipoprotein E4, and the risk of HIV dementia: the Hawaii Aging with HIV Cohort. J Neuroimmunol. 2004 Dec;157(1-2):197–202. doi: 10.1016/j.jneuroim.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 209.Everall I, Vaida F, Khanlou N, et al. Cliniconeuropathologic correlates of human immunodeficiency virus in the era of antiretroviral therapy. J Neurovirol. 2009 Dec;15(5-6):360–370. doi: 10.3109/13550280903131915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Persidsky Y, Gendelman HE. Mononuclear phagocyte immunity and the neuropathogenesis of HIV-1 infection. J Leukoc Biol. 2003 Nov;74(5):691–701. doi: 10.1189/jlb.0503205. [DOI] [PubMed] [Google Scholar]
- 211.Thompson PM, Dutton RA, Hayashi KM, et al. 3D mapping of ventricular and corpus callosum abnormalities in HIV/AIDS. Neuroimage. 2006 May 15;31(1):12–23. doi: 10.1016/j.neuroimage.2005.11.043. [DOI] [PubMed] [Google Scholar]
- 212.Chiang MC, Dutton RA, Hayashi KM, et al. 3D pattern of brain atrophy in HIV/AIDS visualized using tensor-based morphometry. Neuroimage. 2007 Jan 1;34(1):44–60. doi: 10.1016/j.neuroimage.2006.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Thompson PM, Dutton RA, Hayashi KM, et al. Thinning of the cerebral cortex visualized in HIV/AIDS reflects CD4+ T lymphocyte decline. Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15647–15652. doi: 10.1073/pnas.0502548102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chang L, Lee PL, Yiannoutsos CT, et al. A multicenter in vivo proton-MRS study of HIV-associated dementia and its relationship to age. Neuroimage. 2004 Dec;23(4):1336–1347. doi: 10.1016/j.neuroimage.2004.07.067. [DOI] [PubMed] [Google Scholar]
- 215.Ances BM, Roc AC, Wang J, et al. Caudate blood flow and volume are reduced in HIV+ neurocognitively impaired patients. Neurology. 2006 Mar 28;66(6):862–866. doi: 10.1212/01.wnl.0000203524.57993.e2. [DOI] [PubMed] [Google Scholar]
- 216.Schifitto G, Zhong J, Gill D, et al. Lithium therapy for human immunodeficiency virus type 1-associated neurocognitive impairment. J Neurovirol. 2009 Apr;15(2):176–186. doi: 10.1080/13550280902758973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Masliah E, Heaton RK, Marcotte TD, et al. Dendritic injury is a pathological substrate for human immunodeficiency virus-related cognitive disorders. HNRC Group. The HIV Neurobehavioral Research Center. Ann Neurol. 1997 Dec;42(6):963–972. doi: 10.1002/ana.410420618. [DOI] [PubMed] [Google Scholar]
- 218.Achim CL, Adame A, Dumaop W, Everall IP, Masliah E. Increased accumulation of intraneuronal amyloid beta in HIV-infected patients. J Neuroimmune Pharmacol. 2009 Jun;4(2):190–199. doi: 10.1007/s11481-009-9152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Rempel HC, Pulliam L. HIV-1 Tat inhibits neprilysin and elevates amyloid beta. AIDS. 2005 Jan 28;19(2):127–135. doi: 10.1097/00002030-200501280-00004. [DOI] [PubMed] [Google Scholar]
- 220.Green DA, Masliah E, Vinters HV, Beizai P, Moore DJ, Achim CL. Brain deposition of beta-amyloid is a common pathologic feature in HIV positive patients. AIDS. 2005 Mar 4;19(4):407–411. doi: 10.1097/01.aids.0000161770.06158.5c. [DOI] [PubMed] [Google Scholar]
- 221.Esiri MM, Biddolph SC, Morris CS. Prevalence of Alzheimer plaques in AIDS. J Neurol Neurosurg Psychiatry. 1998 Jul;65(1):29–33. doi: 10.1136/jnnp.65.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Nath A, Hauser KF, Wojna V, et al. Molecular basis for interactions of HIV and drugs of abuse. J Acquir Immune Defic Syndr. 2002 Oct 1;31(Suppl 2):S62–69. doi: 10.1097/00126334-200210012-00006. [DOI] [PubMed] [Google Scholar]
- 223.Garden GA. Microglia in human immunodeficiency virus-associated neurodegeneration. Glia. 2002 Nov;40(2):240–251. doi: 10.1002/glia.10155. [DOI] [PubMed] [Google Scholar]
- 224.Bell JE, Brettle RP, Chiswick A, Simmonds P. HIV encephalitis, proviral load and dementia in drug users and homosexuals with AIDS. Effect of neocortical involvement. Brain. 1998 Nov;121(Pt 11):2043–2052. doi: 10.1093/brain/121.11.2043. [DOI] [PubMed] [Google Scholar]
- 225.Everall IP, Hansen LA, Masliah E. The shifting patterns of HIV encephalitis neuropathology. Neurotox Res. 2005 Oct;8(1-2):51–61. doi: 10.1007/BF03033819. [DOI] [PubMed] [Google Scholar]
- 226.Boven LA, Middel J, Breij EC, et al. Interactions between HIV-infected monocyte-derived macrophages and human brain microvascular endothelial cells result in increased expression of CC chemokines. J Neurovirol. 2000 Oct;6(5):382–389. doi: 10.3109/13550280009018302. [DOI] [PubMed] [Google Scholar]
- 227.Mortimer JA, Snowdon DA, Markesbery WR. The effect of APOE-epsilon4 on dementia is mediated by Alzheimer neuropathology. Alzheimer Dis Assoc Disord. 2009 Apr-Jun;23(2):152–157. doi: 10.1097/wad.0b013e318190a855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bennett DA, Schneider JA, Wilson RS, Bienias JL, Berry-Kravis E, Arnold SE. Amyloid mediates the association of apolipoprotein E e4 allele to cognitive function in older people. J Neurol Neurosurg Psychiatry. 2005 Sep;76(9):1194–1199. doi: 10.1136/jnnp.2004.054445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Shulman JM, Chibnik LB, Aubin C, Schneider JA, Bennett DA, De Jager PL. Intermediate phenotypes identify divergent pathways to Alzheimer’s disease. PLoS One. 5(6):e11244. doi: 10.1371/journal.pone.0011244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Bennett DA, De Jager PL, Leurgans SE, Schneider JA. Neuropathologic intermediate phenotypes enhance association to Alzheimer susceptibility alleles. Neurology. 2009 Apr 28;72(17):1495–1503. doi: 10.1212/WNL.0b013e3181a2e87d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sato-Matsumura KC, Berger J, Hainfellner JA, Mazal P, Budka H. Development of HIV encephalitis in AIDS and TNF-alpha regulatory elements. J Neuroimmunol. 1998 Nov 2;91(1-2):89–92. doi: 10.1016/s0165-5728(98)00161-1. [DOI] [PubMed] [Google Scholar]
- 232.Levine AJ, Achim C, Singer EJ, Masliah E, Sinsheimer J, Soontornniyomkij V, Moore DJ. Establishing a Link Between Host Genotype and HIV-Related Neuropathology – A Novel Approach to Understanding Neuropathogenesis. International Neuropsychological Society; Kona, Hawaii. Feb 8, 2012. [Google Scholar]
- 233.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 234.Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 2008;4(8):e1000117. doi: 10.1371/journal.pcbi.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004 Feb;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 236.Voineagu I, Wang X, Johnston P, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011 Jun 16;474(7351):380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Shirasaki DI, Greiner ER, Al-Ramahi I, et al. Network organization of the huntingtin proteomic interactome in mammalian brain. Neuron. 2012 Jul 12;75(1):41–57. doi: 10.1016/j.neuron.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Horvath S, Zhang Y, Langfelder P, et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012 Oct 3;13(10):R97. doi: 10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Song L, Langfelder P, Horvath S. Comparison of co-expression measures: mutual information, correlation, and model based indices. BMC Bioinformatics. 2012;13:328. doi: 10.1186/1471-2105-13-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Langfelder P, Mischel PS, Horvath S. When is hub gene selection better than standard meta-analysis? PLoS One. 2013;8(4):e61505. doi: 10.1371/journal.pone.0061505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Holzinger ER, Hulgan T, Ellis RJ, et al. Mitochondrial DNA variation and HIV-associated sensory neuropathy in CHARTER. J Neurovirol. 2012 Dec;18(6):511–520. doi: 10.1007/s13365-012-0133-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Canter JA, Haas DW, Kallianpur AR, et al. The mitochondrial pharmacogenomics of haplogroup T: MTND2*LHON4917G and antiretroviral therapy-associated peripheral neuropathy. The pharmacogenomics journal. 2008 Feb;8(1):71–77. doi: 10.1038/sj.tpj.6500470. [DOI] [PubMed] [Google Scholar]
- 243.Kamerman PR, Wadley AL, Cherry CL. HIV-associated sensory neuropathy: risk factors and genetics. Current pain and headache reports. 2012 Jun;16(3):226–236. doi: 10.1007/s11916-012-0257-z. [DOI] [PubMed] [Google Scholar]