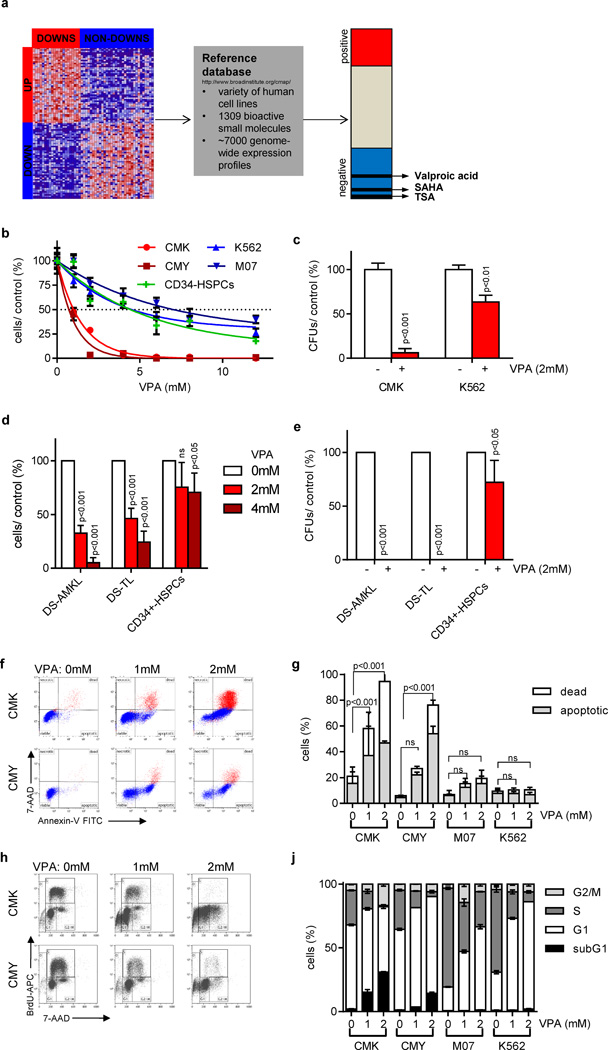
Figure 1. Gene expression-based chemical genomics identifies Down syndrome leukemia to be highly sensitive against HDACis.
(a) Scheme illustrating the setup of the gene expression-based chemical genomic screening, which identified VPA, TSA and SAHA to reverse the DS-AMKL gene signature (17). (b) Number of cells 48h after incubation with the indicated concentrations of VPA relative to the untreated control (=100%). (c) Number of CFUs per 5×102 plated CMK and K562 cells with and without VPA (2mM) relative to the untreated control (=100%). (d) Number of blasts from patients with DS-AMKL (n=3), DS-TL (n=4) and of CD34+-HSPCs (n=2) grown in liquid culture 48h after addition of VPA (2 and 4mM) in relation to the untreated control (white bar; =100%). (e) Number of CFUs of DS-AMKL (n=2) and DS-TL (n=3) blasts as well as CD34+-HSPCs (n=2) in the presence of VPA (2mM) in relation to the untreated control cells (=100%). (d–e) Data are presented as mean±s.d. (f–g) Representative dot plots of Annexin V/ 7-AAD apoptotic assay (f) and diagram showing the percentage of Annexin V+/ 7-AAD- apoptotic and 7-AAD+ dead cells (g) 48h after addition of VPA in comparison to the untreated control cells. (h–j) Representative dot plots (h) and diagram (j) of BrdU/ 7-AAD cell cycle analysis of cells 48h after addition of VPA in comparison to the untreated control cells. (b–c; f–j) All experiments are presented as mean±s.d of at least two independent experiments performed in replicates.