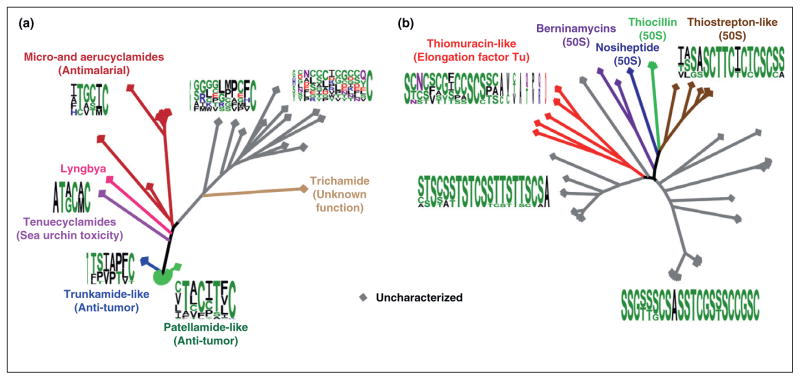
Figure 2.
Phylogenetic analysis of the cyanobactins and thiopeptides. (a) A comparative analysis of the cyanobactins using the publicly available precursor peptide sequences. The core regions of the precursor peptides were used to generate a sequence logo for each clade. In the uncharacterized clades, sequence alignment was used to hypothesize the protease cleavage sites. The least variance is present in the last position of the core peptide, which is predominantly a cysteine that becomes a thiazol(in)e. (b) Same as above, except the thiopeptide precursor peptides were used for the analysis. The clades were classified to bind the 50S ribosomal subunit or elongation factor Tu based on characterized members. All phylogenetic analyses were conducted using MEGA4 [68] and all sequence logos were produced using WebLogo [69].