Abstract
The water-soluble peridinin–chlorophyll a-proteins (PCPs) are one of the major light harvesting complexes in photosynthetic dinoflagellates. PCP contains the carotenoid peridinin as its primary pigment. In this study, we identified and characterized the PCP protein and the PCP gene organization in Symbiodinium sp. CS-156. The protein molecular mass is 32.7 kDa, revealing that the PCP is of the monomeric form. The intronless PCP genes are organized in tandem arrays. The PCP gene cassette is composed of 1095-bp coding regions and spacers in between. Despite the heterogeneity of PCP gene tandem repeats, we identified a single form of PCP, the sequence of which exactly matches the deduced sequence of PCP gene clone 7 (JQ395030) by LC–MS/MS analysis of tryptic digested PCP, revealing the mature PCP apoprotein is 312 amino acids in length. Pigment analysis showed a peridinin-to-Chl a ratio of 4. The peridinin-to-Chl a Qy energy transfer efficiency is 95% in this complex.
Keywords: Photosynthetic antenna, Peridinin–chlorophyll a-protein, Symbiodinium, Peptide sequencing, Absorption spectroscopy, Fluorescence spectroscopy
1. Introduction
Peridinin–chlorophyll a-proteins (PCPs) are one of the major light harvesting complexes in photosynthetic dinoflagellates [1-3]. PCP is water-soluble and has the blue-green (470 nm to 550 nm) absorbing carotenoid peridinin as its primary pigments. Symbiodinium, the most commonly found endosymbiotic dinoflagellates in symbiosis with corals [4,5], also contain PCPs as their major light harvesting complexes [6-8]. A recent study [9] suggested that Symbiodinium PCP may play an important role in coral bleaching, which results from the loss of Symbiodinium cells from the coral host or the loss of photosynthetic pigments from Symbiodinium [4]. Because coral bleaching always follows severe photoinhibition [9-11], and moderate heat stress can induce the algal photosystem II (PSII) photoinhibition by damaging photosynthetic light harvesting complexes and thylakoid membranes [9,12], it is hypothesized that increased sea surface temperature is one of the major factors that trigger coral bleaching. The study [9] showed that the level of PCP in a cultured thermal-sensitive Symbiodinium strain dropped under light conditions when temperature increased, supporting the temperature-induced coral bleaching theory and suggesting the connection between Symbiodinium PCP and coral bleaching.
However, Symbiodinium PCP is poorly understood in the aspects of structure, spectroscopic properties, energy transfer, etc. In general, dinoflagellate PCP proteins are very diverse. The length, pigment content, sequence and spectroscopic properties of PCP can be distinct among different dinoflagellates or even in an individual dinoflagellate. In the aspect of length, there are two forms of PCPs: one is a homodimer with a monomeric molecular mass of ~15 kDa [13,14], the other is a monomer with a molecular mass of 32–35 kDa [3,13,14].The latter form was hypothesized to be the product of duplication and subsequent fusion of the PCP gene encoding the former form [8,15]. Two monomers of the homodimeric PCP probably form a dimer, whose structure could be very similar to that of the monomeric PCP monomer [3,16].In Amphidinium carterae, three copies of the monomeric PCP associate to form a trimeric complex [3,17]. The species specificity of the PCP quaternary structure has been reported [14]. Both monomeric and homodimeric forms were detected in Symbiodinium microadriaticum, while Symbiodinium kawagutii and Symbiodinium pilosum only possess a single form of PCP, monomeric and homodimeric forms, respectively [14]. In the aspect of pigment content of PCP, the molar ratio of peridinin and Chl a varies: the main form PCP complex (MFPCP) from A. carterae has the pigment composition of 8:2 [3]; the high-salt PCP (HSPCP) from the same species, eluted at higher salt concentration from an ion exchange column, has 6 peridinins and 2 Chl a molecules in one monomer [18]; Haxo et al. reported a 9:2 ratio for the PCP from Amphidinium carterae Plymouth 450 [1];PCP from Alexandrium cohorticula may consist of 12 peridinins and 2 Chl a molecules [19].
The PCP protein sequence is not very conserved, leading to distinct pigment binding environments, which determine the spectroscopic properties of a protein [20]. The MFPCP and HSPCP from A. carterae have a sequence similarity of ~30%, resulting in peridinin loss and chlorophyll phytol chains rearrangement in HSPCP [18,20]. Besides, PCP isoforms with varying isoelectric points (pIs) [1,2,5,14,15] were observed in dinoflagellates. The major isoforms tend to be species-specific. A. carterae and S. microadriaticum mainly produce basic PCPs of pI 7.5 and pI 7.2–7.7, respectively [1,14], while Gonyaulax polyedra PCPs are mostly acidic of pI around 6 [15]. Small spectroscopic variations of PCP isoforms were reported in S. microadriaticum: the Chl a Qy absorption maxima varied from 669.8 nm to 673.4 nm and the corresponding fluorescence emission maxima were from 673.0 nm to 676.5 nm [14].
In order to understand the source and significance of PCP isoforms, PCP protein structures and PCP genes have been explored since 1990s. Among different PCPs, the structures of MFPCP and HSPCP from A. carterae have been resolved to 2.0 Å and 2.1 Å, respectively [3,18]. A higher resolution of 1.5 Å was recently resolved in refolded PCP [21]. The X-ray crystal structure of MFPCP has revealed a trimer, in which each subunit folds in a twofold pseudosymmetry (a monomer has two pseudo-identical domains), holding 8 peridinins and 2 Chl a molecules. On the other hand, HSPCP crystallized as a monomer, which contains 6 peridinins and 2 Chl a molecules. The high degree of structural similarity was observed, although sequence variations led to the differences in the level of the protein scaffold, pigment composition, and interaction between the pigments and their binding environment [18]. The sequence analysis of PCP genes and transcripts based on the data from A. carterae [22], G. polyedra [15], Heterocapsa pygmaea [16,23] and Symbiodinium sp. [8,13,24] revealed that PCP genes are nuclear-encoded, intronless and exist in tandem arrays. PCP gene arrays consist of coding regions and the spacers in between. The lengths of coding regions and spacers, as well as the copy number of PCP genes, are species-specific. The sequences of coding regions in PCP gene arrays are distinct among different dinoflagellate species and even in one individual dinoflagellate. Along with the observation of multiple PCP isoforms with varying pIs, the PCP gene heterogeneity is believed to be the source of the isoforms. However, except for two different forms of PCP proteins (MFPCP and HSPCP from A. carterae), which have been detected and determined at the protein sequence and structure level [3,16,18], there has been no report successfully relating cDNA or gDNA sequences to their corresponding protein sequences because of the complexity of PCP genes and the possible protein expression preferences.
In this study, we identified the PCP gene family in Symbiodinium sp. CS-156 [25], confirming the genetic diversity of PCP genes and the existence of PCP tandem arrays. We further related PCP gene sequence to its corresponding protein sequence and compared the spectroscopic properties of Symbiodinium PCP with those of Amphidinium PCP.
2. Materials and methods
2.1. Algal culture and PCP purification
Symbiodinium sp. CS-156 cells were cultured in f/2 media under a 14 h:10 h cycle of light:dark at 25 °C. Illumination was provided by a white color fluorescent lamp at an intensity of 80 μmol photon·m−2·s−1. The culture in late exponential phase was harvested by centrifugation at 8000 g for 10 min at 4 °C. Cells were resuspended in 50 mM tricine 20 mM KCI (pH 7.5), and broken by three passes through a French pressure cell at 8.3×107 Pa [22]. Cell debris and unbroken cells were removed by centrifugation at 20,000 g for 1 h. Solid ammonium sulfate was added to the resulting supernatant to achieve 70% saturation, followed by centrifugation at 8000 g for 15 min. The resulting supernatant was dialyzed against 20 μM Tris–HCl (pH 8.0), concentrated, filtered through a 0.2 μm filter, and applied to a HiLoad™ Superdex™ 200 prep grade column. The PCP fraction eluted by 20 mM Tris–HCl (pH 8.0), was then applied to a HiTrap™ Q Sepharose™ HP column and eluted with a linear gradient of NaCl from 0 to 0.5 M in the same buffer. SDS-PAGE was performed to confirm the size and purity of PCP [26].
2.2. LC/MS analysis of PCP
The PCP protein sample was analyzed by a Synapt G2 Q-IM-TOF mass spectrometer coupled with a NanoAcuity UPLC (Waters Inc., Milford, MA) as previously described [27] with minor modifications. The protein sample was directly loaded onto a home-packed C18 column (Magic, 0.075 mm × 50 mm, 5 μm, 120 Å, Michrom Bioresources, Inc., Auburn, CA) by a six-port injection valve (IDEX Health & Science, Oak Harbor, WA). The gradient was delivered by NanoAcuity UPLC (0–2 min, 5% solvent B; 2–15 min, 5–95% solvent B. Solvent A: water, 0.1% formic acid; Solvent B: acetonitrile, 0.1% formic acid) at flow rate 1 μL/min. The protein spectrum was acquired at sensitive mode (“v” mode) with the capillary voltage of 1.8 kV, cone voltage of 30 V and source temperature of 100 °C.
2.3. Peptide sequencing of tryptic digested PCP
The Coomassie blue-stained band was excised and in-gel digested with trypsin as previously described [28] with minor modifications. After dithiothreitol reduction and iodoacetamide alkylation, 20 μg/mL trypsin (Sigma) in 36 mM NH4HCO3, 8.1% acetonitrile, and 0.1 mM HCl was added to the gel pieces. After incubation at 37 °C for 12 h and centrifugation, the supernatant was collected, and the gel pellet was extracted with 1% trifluoroacetic acid in 60% acetonitrile for 30 min. Then supernatants were combined and dried by vacufuge. The trypsin digested PCP sample was reconstituted in water with 0.1% formic acid (25 μL). The sample was analyzed by LC–MS/MS using two instruments, a Waters Synapt G2 Q-IM-TOF and a Thermo LTQ Orbitrap (Thermo-Scientific, San Jose, CA). The data dependent mode was used in the LC–MS/MS experiment at Orbitrap as previously described [29] with minor modifications. The MSE mode was used in the LC–MS/MS experiment on the Synapt G2 [30]. The tryptic digested PCP sample (5 μL) was loaded onto a homemade silica capillary column that was custom packed with C18 reverse phase material (Magic, 0.075 mm × 150 mm, 5 μm, 120 Å, Michrom Bioresources, Inc., Auburn, CA). The gradient was supplied by a Waters NanoAquity UPLC and run from 5% solvent B (acetonitrile, 0.1% formic acid) to 50% solvent B over 60 min, then to 95% solvent B for 2 min at 400 nL/min flowed by a re-equilibration step with 100% solvent A (water, 0.1% formic acid). The flow was directed by a nanospray source (Waters, Inc., Milford, MA). In the MSE continuum mode, ions were dissociated in the trap region by ramping the trap collision energy from 14–40 V. Spectra from 50 to 2000 m/z were acquired with scan time 1 s for 70 min in positive sensitivity mode. The MSE raw data were directly submitted to the ProteinLynx Global Server (V2.5, Waters Inc., Milford, MA) to search against the NCBI and UniProt database.
2.4. PCP gene identification
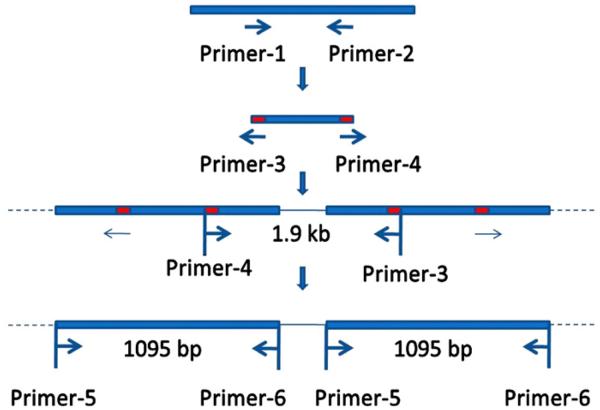
Symbiodinium genomic DNA was extracted by Qiagen® DNeasy Plant Mini Kit. PCP genes were cloned by a method previously developed for Symbiodinium with some modifications (Fig. 1) [24]. Briefly, primer-1 and primer-2 amplified a ~200 bp fragment of PCP gene; primer-3 and primer-4, designed according to the sequence of the ~200 bp fragment [24], amplified a ~1.9 kb fragment containing partial PCP genes and a linker; primer-5 and primer-6 were designed according to the possible start and stop codon sites in the 1.9 kb fragment. The PCR products amplified by primer-5 and primer-6 were cloned and analyzed. Primers used for PCR are listed in Table 1. The purified PCR products were cloned and sequenced. Sequences were analyzed by NCBI BLAST and Vector NTI®.
Fig. 1.
The strategy to identify PCP genes in Symbiodinium sp. CS-156-b1 [24].
Table 1.
Primers and the annealing conditions.
| Primer name | Primer sequence (5′–3′) | Annealing |
|---|---|---|
| Primer-1 | AAGAATTCGAAGGACGCAGCAGAAGC [24] | 30 s at 52 °C |
| Primer-2 | CAGAATTCOTCATGTACGCTGGCAC [24] | |
| Primer-3 | TCGGTCCCCAAAGCAAAGGTCA [24] | 30 s at 55 °C |
| Primer-4 | CATTCACGGCATCCCAGTCAGC [24] | |
| Primer-5 | TGGTGCGTGGAGCAAGGAAA | 30 s at 55 °C |
| Primer-6 | TTCACCTTCAGCGCTGGGAA |
2.5. Spectroscopic characterization
2.5.1. Steady-state absorption spectroscopy
The PCP complex was suspended in 10 mM Tris–HCl (pH 8.0) containing 60% glycerol. Steady-state absorption spectra at both room temperature and 77 K were recorded using a Perkin-Elmer Lambda 950 UV–vis spectrophotometer. The OptistatDN (Oxford Instruments, UK) liquid nitrogen cryostat was used to cool the protein sample to 77 K.
2.5.2. Fluorescence spectroscopy
The PCP complex was suspended in the same buffer as described in 0. The fluorescence emission and excitation spectra at both room temperature and 77 K were obtained using a Photon Technology International fluorometer. The cryostat described in 0 was used in 77 K spectra measurement. Emission and excitation monochromator slits were set to a bandpass of 4 nm. The fluorescence excitation spectrum was corrected using a calibrated reference photodiode. The fluorescence excitation spectrum shown is the average of 10 individual spectra.
2.6. Pigment determination
The purified PCP complex in 20 mM Tris–HCl (pH 8.0) was dried in darkness by vacufuge. 200 μL methanol was added to the dry PCP, followed by brief vortexing and centrifugation. The resulting supernatant was collected, and the methanol extraction was repeated 4 times. All the supernatants were combined, then well mixed. Peridinin and Chl a were extracted by methanol from Symbiodinium whole cells, and separated by Agilent 1100 HPLC on a Zorbax Eclipse XDB-C18 reverse phase column (250 mm × 4.6 mm) [31]. Peridinin and Chl a fractions eluted from the column were dried in darkness by vacufuge and redissolved in methanol. The steady-state absorption spectra of the PCP methanol extract, peridinin and Chl a were recorded using a Perkin-Elmer Lambda 950 UV–vis spectrophotometer. The extinction coefficients used for the pigments ratio calculation were:
Chl a (methanol): ε665 nm=71.43 L·mmol−1·cm−1 [32]
Peridinin (methanol): ε469 nm=85.80 L·mmol−1·cm−1 [33,34]
The molar ratio was calculated according to the Beer-Lambert Law:
A=ε·c·l (A: absorbance, c: molar concentration, l: optical path length).
3. Results and discussion
3.1. PCP identification
The PCP of Symbiodinium sp. CS-156 was purified by ammonium sulfate precipitation, gel filtration chromatography and ion exchange chromatography. The purity and size of PCP were examined by SDS-PAGE and LC/MS (Fig. 2). SDS-PAGE revealed a single band at a mass of 33 kDa, which was determined to be 32.7 kDa by LC/MS. No protein peak around 14–16 kDa was observed by LC/MS, indicating that the PCP of Symbiodinium sp. CS-156 is of the monomeric form. Peptide sequencing identified multiple peptides that are part of PCP putative preproteins in UniProt database. The one with most hits is a PCP (the UniProtKB/Swiss-Prot entry: P51874) from Symbiodinium sp., which is a cDNA-translated protein sequence [8]. This PCP preprotein contains a transit peptide, and the predicted mature protein is composed of 2 almost identical repeat units. The theoretical molecular weight of the mature P51874 PCP gene product is 32585.18 Da.
Fig. 2.
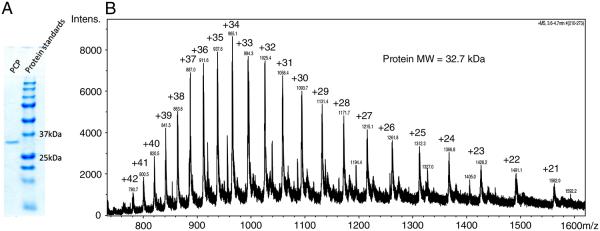
The identification of PCP by SDS-PAGE (A) and LC/MS (B). SDS-PAGE revealed a single band at a mass of about 33 kDa, which was determined to be 32.7 kDa by LC/MS.
PCP genes were cloned and sequenced. Among 11 PCP gene clones (see Supplementary Data), only 2 of them are identical in nucleotide sequence (GenBank ID: JQ395029 to JQ395038), indicating that a variation of PCP genes exist at the gene level as was found for free-living dinoflagellates in previous studies [23,24]. The comparison of 11 genomic DNA sequences with Symbiodinium PCP cDNA sequences in previous studies [8,13,24], showed no evidence of introns. Each clone encodes a 365 amino acid preprotein, which corresponds to a 1095 bp coding region of the PCP gene. The predicted preproteins have identities ranging from 96.4% to 100% when aligned with each other. The observed identities are close to the reported values based on the analysis of PCP cDNA clones [24], which are from 96.2% to 99.7% identical. The small variations of PCP genes increase the genetic diversity, introducing small differences in deduced preprotein mass (by ExPASy Compute pI/Mw tool, from 32633.52 Da to 32868.63 Da) but relatively large differences in their predicted isoelectric points (pIs from 5.34 to 7.91).
Sequencing the 1.9 kb clones shows that each clone contains open reading frames that can be translated to PCP at both 5′ and 3′ ends, and a spacer region in the middle of the two ORFs. This confirms that PCP genes of Symbiodinium sp. CS-156 are organized in tandem arrays as was found in other Symbiodinium strains [24]. As anticipated, the spacer sequences are highly varied, containing several insertions and deletions (data not shown).
In the LC–MS/MS peptide sequencing experiment, peptides from PCP protein were fragmented in the mass spectrometer by collision-induced dissociation. The protonated peptides fragment along the peptide backbone to form b and y production ions. The peptide sequences were elucidated from the match between in-silico digested protein sequence and sequence product ions from experiment [35]. All detected peptides were processed by MassMatrix Database Search Engine [36,37]. The comparison of the translated PCP sequences with peptide sequencing by LC–MS/MS, revealed that the mature protein starts at Asp53. Protein sequence alignment of five PCP proteins (which exist at the protein level, UniProt entries: P51873 [22], P51874 [8], P51889 [19], P80483 [22] and P80484 [3]) and translated sequences in this study shows that all mature PCPs begin at Asp53, and except for the HSPCP of A. carterae, other proteins share a sequence of DEIGDAAK at the N terminus. The search results also show that the sequence of the trypsin digested sample exactly matches the deduced sequence of clone 7(Fig. 3). The calculated MW of the putative PCP mature protein deduced from clone L4 is 32.8 kDa, which is in agreement with the MW of 32.7 kDa obtained by LC/MS. Compared to other protein sequencing methods, e.g. Edman degradation [8,13] and protein microsequencing [15], the LC–MS/MS protein sequencing obtained a full coverage, which is the highest to our knowledge for any PCP.
Fig. 3.
PCP protein sequence identification. The listed sequence is the predicted amino acid sequence of a PCP gene clone (Clone 7, JQ395030). LC/MSMS experiment by UPLC-QTOF (Waters, Synapt G2) identified multiple peptides, assembly of all of which matches the predicted sequence of clone 7. The transit peptide is in green; the solid-underlined sequence in green is the possible thylakoid-targeting domain; the two boxes indicate the possible Chl a molecules binding sites; the dotted-underlined sequences are the possible peridinin binding sites.
We explored more characteristics of PCP based on DNA and protein sequence. As expected, the first half of the mature PCP apoprotein (Asp53–Ala208) has 61% identity with the second half (Pro209–Gln364). An identity of 54% was observed in previous studies [8,15]. Along with the two-fold symmetry of PCP X-ray crystal structure [3,18], our finding supports the hypothesis that the monomeric form of PCPs arose by duplication and fusion of gene(s) encoding the homodimeric form [8,15]. The transit sequence (under-lined sequence in green in Fig. 3) adjacent to the mature PCP sequence has two distinct hydrophobic regions when analyzed by ExPASy ProtScale. This transit peptide (TP) shares an identity of 43% with the G. polyedra PCP transit peptide [15,38], which contains an additional hydroxylated amino acids (S/T)-rich region flanked with two distinct hydrophobic regions. The first region could guide the nuclear-encoded protein translocation from cytosol to chloroplast, and then be cleaved off, generating an intermediate preprotein; the second region could lead the intermediate to thylakoids where a thylakoidal processing peptidase (TPP) recognizes the A–X–A motif in TP, cleaves TP off and generates the mature apoprotein [39-41]. The second hydrophobic regions of both G. polyedra and Symbiodinium sp. TPs are rich in alanine, a typical feature of several TPs of nuclear-encoded thylakoid proteins [40]. The TP sequence analysis is in agreement with the co-crystallization of PCP and DGDG [3], which is mostly found in the inner thylakoid membrane [42], supporting that PCP is located in the thylakoid lumen [8,15].
To our knowledge, this is the first report that relates PCP gene sequence to its corresponding protein sequence in the genus Symbiodinium. Previous attempts only included cDNA sequence [16,23], or a partial PCP protein sequence [19], or a very small portion of PCP protein sequence that corresponds to cDNA [8,13,15], except for the MFPCP and HSPCP from A. carterae, whose crystal structures have been determined [3,18,22]. Only three groups have reported the descriptions of the PCP gene organization in Symbiodinium [8,13,24], but none of them was able to determine the PCP sequence at both gene and protein level. The sequencing and molecular experiments show that the major form of PCP in Symbiodinium sp. CS-156 is monomeric, and intronless PCP genes are organized in tandem arrays. The arrangement of PCP genes in this organism is very similar to that of other dinoflagellates, although the DNA and protein sequences are different from those of others. The occurrence of heterogeneity in the PCP gene tandem repeats is also observed. The PCP gene heterogeneity was believed to be inconsistent with concerted evolution [24], which leads to the sequences of related genes to co-evolve over some period of time. However, although the variations can be detected at both mRNA [22,24] and protein level [1,2,6,13,14], the number of major isoforms in an individual organism seems to be very limited: an isoform with a single pI or isoforms with subtle pI differences always overwhelm other PCP species. This situation could also be applied to Symbiodinium sp. CS-156: we have found that there are some other isoforms expressed, but that the one characterized in this study is the main one (data not shown). It is noted that the hypothesis of the existence of major isoform(s) does not conflict with the fact that both monomeric and homodimeric forms were detected in S. microadriaticum [14], in which major isoforms have pIs of 7.2, 7.3, 7.6 and 7.7. Since each fraction with the same pI was able to separate into two PCP species with distinct sizes but the same spectroscopic properties, the monomeric form could have arisen by duplication and fusion of gene(s) encoding the homodimeric form: the two PCPs of different lengths but the same pI could be the products of the same gene. In sum, there could be some posttranscriptional mechanism that leads to the mature apoPCP expression preference. At this point, the PCP expression pattern appears to be consistent with the concerted evolution model.
3.2. Absorption spectroscopy
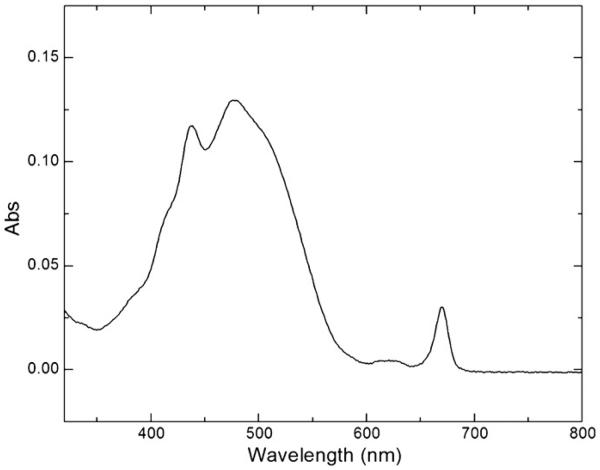
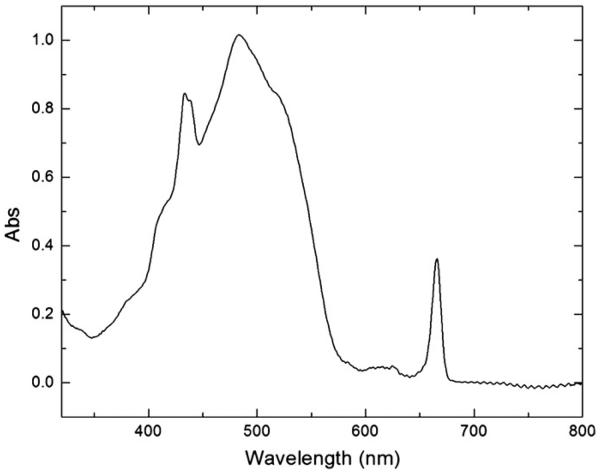
The absorption spectrum of PCP measured at room temperature is shown in Fig. 4. The spectrum consists of a broad band between 400 and 550 nm, two shoulders at 420 nm and 520 nm, and three peaks at 438 nm, 476 nm and 670 nm. The peridinin S0–S2 transition results in the dominant broad band between 400 and 550 nm with the peridinin absorption maximum at 476 nm. The Chl a Soret band, which overlaps with the broad peridinin band, is at 438 nm. The Chl a Qy band peaks at 670 nm, while the weak Qx band is overlapped by the peridinin S0–S2 transition band. The absorption spectrum taken at 77 K (Fig. 5) has a similar shape, except that the Chl a Soret band is split into two peaks with maxima at 433 nm and 438 nm, indicating the different protein environments of two Chl a molecules. To locate the possible Chl a binding sites in the Symbiodinium PCP, the Symbiodinium PCP putative protein sequence (as listed in Fig. 3) was aligned with the A. carterae PCP (PDB ID: 1PPR), in which His66 interacts with Chl a through a water molecule in the first half of the mature protein [43], and Leu254 along with His229 are part of the environment of the COOH-terminal chlorophyll [3] in the second half of the mature protein. The two boxes in Fig. 3 are very likely to bind Chl a molecules in Symbiodinium PCP because these two fragments have the same amino acid sequences with the PCP (PDB ID: 1PPR) Chl a binding regions (the region around His66 and the region around Leu254 and His229). Region 1 (AAEAHHKAIGSISGPNGVTSRADWD) and region 2 (LKAAAEAHHKAIGSIDA) share a sequence of AAEAHHKAIGSI, but have the rest of the amino acid residues with distinct properties, resulting in different protein environments that can be detected in 77 K absorption spectrum.
Fig. 4.
The absorption spectrum of PCP at room temperature.
Fig. 5.
The absorption spectrum of PCP at 77 K.
In addition, at 77 K, the peridinin absorption maximum is red-shifted from 476 nm to 483 nm, while the Chl a Qy band is blue-shifted from 670 nm to 666 nm. Compared with the RT absorption spectrum, the 77 K spectrum has more distinct shoulders at 420 nm and 520 nm and more resolved Qy 0–1 vibronic and/or Qx transition band between 600 nm and 650 nm [44]. This is because the lower temperature reduces the randomness of orientation of protein molecules in the system, thus providing more information.
In general, the steady-state absorption spectra of Symbiodinium PCP at both temperatures are very similar to those of A. carterae PCP in the aspects of shape and locations of peaks and shoulders except for some minor differences (e.g. the ratios of peridinin absorption maximum to Chl a Qy peak at both temperature are slightly different). The protein sequence analysis revealed the conservative binding sites for peridinin and Chl a molecules in Symbiodinium PCP (Fig. 3, two boxes and two dotted-underlined sequences are the possible binding pockets for Chl a and peridinin molecules, respectively) [3,43,45]. Symbiodinium PCP and A. carterae PCP share AAEAHHKAIGSISGPNGVTSRADWD and LKAAAEAHHKAIGSIDA for Chl a binding, as well as VNAALGRV and VNAALGR for peridinin binding, leading to the almost identical absorption spectra, which are dependent on the protein environments of the pigments.
3.3. Fluorescence spectroscopy
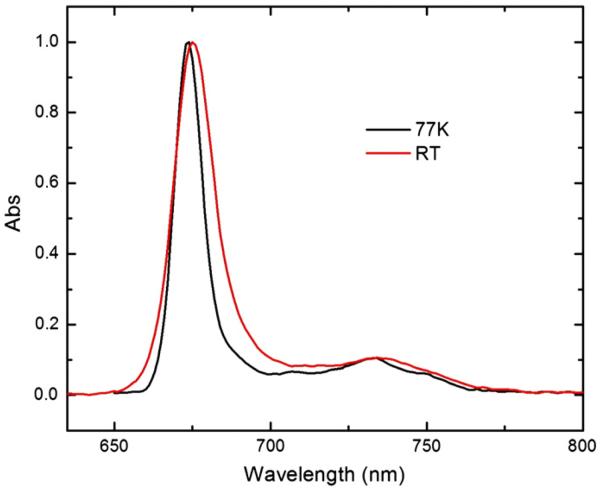
The fluorescence emission spectra of PCP were taken at room temperature and 77 K (Fig. 6). The two spectra exhibit a similar shape, with a peak maximum at 675 nm and 674 nm, respectively, and a broad vibronic band between 700 and 770 nm, centered around 735 nm. The 77 K spectrum is narrower than the RT spectrum: the former has a full width at half-maximum (fwhm) value of 10 nm, while the latter has a value of 15 nm.
Fig. 6.
The fluorescence emission spectra of PCP at room temperature (red) and 77 K (black).
Symbiodinium PCP fluorescence emission spectra at both temperatures are very similar to A. carterae MFPCP and HSPCP spectra, as well as the spectra of Chl a in 2-MTHF [20] (Fig. 6). All the spectra are typical of monomeric Chl species [20]. However, the fluorescence emission maxima of 3 PCPs are different (Table 2). The Symbiodinium PCP spectrum maximum only blue-shifts by 1 nm upon cooling to 77 K, as opposed to 4 nm of the MFPCP spectrum maximum. Although the HSPCP spectrum red-shifts by 1 nm, the spectral maxima are closest to those of Symbiodinium PCP.
Table 2.
The comparison of Symbiodinium PCP, Amphidinium carterae MFPCP and HSPCP [20], and Chl a (in 2-MTHF) [20] fluorescence emission spectra at room temperature and 77 K.
| Peak maximum at RT | Peak maximum at 77 K | |
|---|---|---|
| Symbiodinium PCP | 675 nm | 674 nm |
| Amphidinium carterae MFPCP | 673 nm | 669 nm |
| Amphidinium carterae HSPCP | 674 nm | 675 nm |
| Chl a in 2-MTHF | 668 nm | 671 nm |
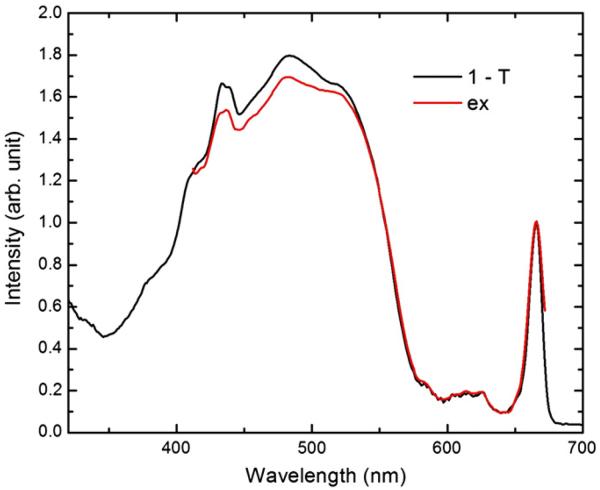
The fluorescence excitation spectrum taken at 77 K was overlaid with the corresponding 1-T spectrum, as shown in Fig. 7. The fluorescence excitation spectrum was normalized with the 77 K 1-T spectrum Qy band. The height difference between the peridinin absorption peaks of both spectra indicates the peridinin-to-Chl a energy transfer loss. The calculated energy transfer efficiency is 95%, which is very close to the reported peridinin-to-Chl a energy transfer efficiencies [20].
Fig. 7.
The overlay of fluorescence excitation (ex) spectrum at 77 K (red) with the corresponding 1-T spectrum (black). The fluorescence excitation spectrum was normalized at the 77 K 1-T spectrum Qy band. The calculated peridinin-to-Chl a energy transfer efficiency is 95%.
3.4. Stoichiometry of pigments in PCP
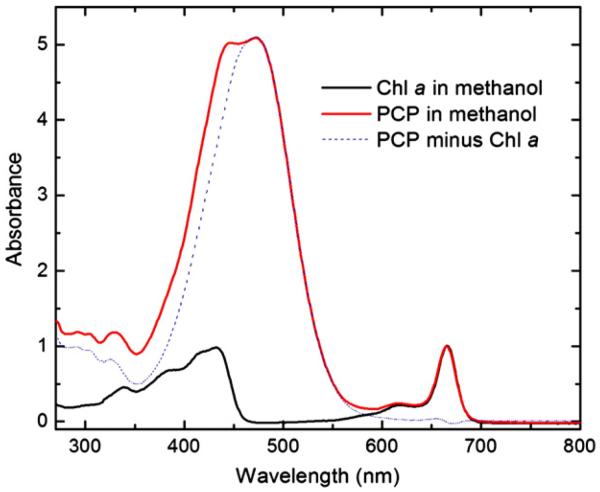
The molar ratio of peridinin to Chl a was determined by measuring the steady-state absorbance of the PCP methanol extract. The UV–vis absorption spectra of peridinin and Chl a fractions eluted from HLPC were confirmed by comparison with literature values [31,46] (data not shown). The spectra of the PCP methanol extract and Chl a in methanol were normalized at the Chl a Qy band. The contribution of Chl a absorbance at 469 nm was deducted from the PCP methanol extract absorbance at the same wavelength (Fig. 8). The calculated value of the molar ratio is 4.07, which is very close to 4. This result along with the spectroscopic properties indicates that Symbiodinium PCP is very likely to share the same structure of A. carterae MFPCP, although the sequence identity is ~83% based on the alignment of the Symbiodinium PCP sequence (listed in Fig. 3) with the A. carterae PCP (PDB ID: 1PPR). The pigment stoichiometry and the conservative pigment binding sites suggest that there is consistently high energy transfer efficiency in dinoflagellate PCPs.
Fig. 8.
The spectra of the PCP methanol extract (solid red) and Chl a (in methanol) (solid black), normalized at Chl a Qy band. The difference between the solid red line and the solid black is indicated by the dotted blue line, which matches the shape of peridinin absorption spectrum. The contribution of Chl a absorbance at 469 nm was deducted from the PCP methanol extract absorbance at the same wavelength.
4. Conclusion
In this study, we identified and characterized PCP and its gene organization in Symbiodinium sp. CS-156. We found that this strain possesses the monomeric form of PCP of 32.7 kDa, encoded by the PCP gene cassette composed of 1095-bp coding regions and spacers in between. Despite the occurrence of the PCP gene heterogeneity, a single form of PCP was identified and sequenced, indicating the possible existence of a PCP major form. The resulting sequence matches the deduced sequence of PCP gene clone 7. The detection of PCP protein and its corresponding gene is first reported in the dinoflagellate Symbiodinium and may provide an important hint for future evolutionary studies. The protein sequence analysis, along with the spectroscopic studies, revealed the conservative pigment binding sites, which may be one of the key factors that determine the consistently high peridinin-to-Chl a energy transfer efficiency in dinoflagellate PCPs.
Supplementary Material
Acknowledgements
The authors would like to thank Drs. David Kramer and Atsuko Kanazawa of Michigan State University for providing the Symbiodinium culture. We would like to thank the NIH NCRR Biomedical Mass Spectrometry Resource at Washington University supported by NIH NCRR Grant P41RR000954, for Orbitrap LC–MS/MS experiments. We would also like to thank Dr. Dariusz Niedzwiedzki and Dr. Haijun Liu for discussions, and Ms. Mindy Prado for helping with culturing. This research is from the Photosynthetic Antenna Research Center (PARC), an Energy Frontier Research Center funded by the DOE, Office of Science, Office of Basic Energy Sciences under Award Number DE-SC 0001035.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at doi:10.1016/j.bbabio.2012.03.027.
References
- 1.Haxo FT, Kycia JH, Somers GF, Bennett A, Siegelman HW. Peridinin–chlorophyll a proteins of the dinoflagellate Amphidinium carterae (Plymouth 450) Plant Physiol. 1976;57:297–303. doi: 10.1104/pp.57.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prezelin BB, Haxo FT. Purification and characterization of peridinin–chlorophyll a proteins from marine dinoflagellates Glenodinium sp. and Gonyaulax polyedra. Planta. 1976;128:133–141. doi: 10.1007/BF00390314. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann E, Wrench PM, Sharples FP, Hiller RG, Welte W, Diederichs K. Structural basis of light harvesting by carotenoids: peridinin–chlorophyll-protein from Amphidinium carterae. Science. 1996;272:1788–1791. doi: 10.1126/science.272.5269.1788. [DOI] [PubMed] [Google Scholar]
- 4.Stat M, Carter D, Hoegh-Guldberg O. The evolutionary history of Symbiodinium and scleractinian hosts — symbiosis, diversity, and the effect of climate change, Perspect. Plant Ecol. 2006;8:23–43. [Google Scholar]
- 5.Trench RK, Blank RJ. Symbiodinium microadriaticum Freudenthal, Symbiodinium goreauii sp. nov., Symbiodinium kawagutii sp. nov. and Symbiodinium pilosum sp. nov.:gymnodinioid dinoflagellate symbionts of marine invertebrates. J. Phycol. 1987;23:469–481. [Google Scholar]
- 6.Chang SS, Trench RK. The isoelectric forms, quaternary structure and amino acid composition of peridinin chlorophyll a proteins from the symbiotic dinoflagellate Symbiodinium microadriaticum Freudenthal. Proc. Royal Soc. Lond. B Biol. Sci. 1984;222:259–271. [Google Scholar]
- 7.Iglesias-Prieto R, Trench RK. Acclimation and adaptation to irradiance in symbiotic dinoflagellates. II. Response of chlorophyll–protein complexes to different photon-flux densities. Mar. Biol. 1997;130:23–33. [Google Scholar]
- 8.Norris BJ, Miller DJ. Nucleotide sequence of a cDNA clone encoding the precursor of the peridinin-chlorophyll a-binding protein from the dinoflagellate Symbiodinium sp. Plant Mol. Biol. 1994;24:673–677. doi: 10.1007/BF00023563. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi S, Whitney S, Itoh S, Maruyama T, Badger M. Heat stress causes inhibition of the de novo synthesis of antenna proteins and photobleaching in cultured Symbiodinium. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4203–4208. doi: 10.1073/pnas.0708554105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warner ME, Fitt WK, Schmidt GW. The effects of elevated temperature on the photosynthetic efficiency of zooxanthellae in hospite from four different species of reef coral: a novel approach. Plant Cell Environ. 1996;19:291–299. [Google Scholar]
- 11.Warner ME, Fitt WK, Schmidt GW. Damage to photosystem II in symbiotic dinoflagellates: a determinant of coral bleaching. Proc. Natl. Acad. Sci. U. S. A. 1999;96:8007–8012. doi: 10.1073/pnas.96.14.8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi S, Whitney SM, Badger MR. Different thermal sensitivity of the repair of photodamaged photosynthetic machinery in cultured Symbiodinium species. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3237–3242. doi: 10.1073/pnas.0808363106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weis VM, Verde EA, Reynolds WS. Characterization of a short form peridinin–chlorophyll-protein (PCP) cDNA and protein from the symbiotic dinoflagellate Symbiodinium muscatineu (dinophyceae) from the sea anemone Anthopleura elegantissima (cnidaria) J. Phycol. 2002;38:157–163. [Google Scholar]
- 14.Iglesiasprieto R, Govind NS, Trench RK. Apoprotein composition and spectroscopic characterization of the water-soluble peridinin chlorophyll alpha-proteins from 3 symbiotic dinoflagellates. Proc. Royal Soc. Lond. B Biol. Sci. 1991;246:275–283. [Google Scholar]
- 15.Le QH, Markovic P, Hastings JW, Jovine RV, Morse D. Structure and organization of the peridinin–chlorophyll a-binding protein gene in Gonyaulax polyedra. Mol. Gen. Genet. 1997;255:595–604. doi: 10.1007/s004380050533. [DOI] [PubMed] [Google Scholar]
- 16.Hiller RG, Crossley LG, Wrench PM, Santucci N, Hofmann E. The 15-kDa forms of the apo-peridinin–chlorophyll a protein (PCP) in dinoflagellates show high identity with the apo-32 kDa PCP forms, and have similar N-terminal leaders and gene arrangements. Mol. Genet. Genomics. 2001;266:254–259. doi: 10.1007/s004380100551. [DOI] [PubMed] [Google Scholar]
- 17.Wormke S, Mackowski S, Schaller A, Brotosudarmo TH, Johanning S, Scheer H, Brauchle C. Single molecule fluorescence of native and refolded peridinin–chlorophyll-protein complexes. J. Fluoresc. 2008;18:611–617. doi: 10.1007/s10895-008-0310-9. [DOI] [PubMed] [Google Scholar]
- 18.Schulte T, Sharples FP, Hiller RG, Hofmann E. X-ray structure of the high-salt form of the peridinin–chlorophyll a-protein from the dinoflagellate Amphidinium carterae: modulation of the spectral properties of pigments by the protein environment. Biochemistry. 2009;48:4466–4475. doi: 10.1021/bi802320q. [DOI] [PubMed] [Google Scholar]
- 19.Ogata T, Kodama M, Nomura S, Kobayashi M, Nozawa T, Katoh T, Mimuro M. A novel peridinin–chlorophyll a protein (PCP) from the marine dinoflagellate Alexandrium cohorticula: a high pigment content and plural spectral forms of peridinin and chlorophyll a. FEBS Lett. 1994;356:367–371. doi: 10.1016/0014-5793(94)01314-4. [DOI] [PubMed] [Google Scholar]
- 20.Ilagan RP, Shima S, Melkozernov A, Lin S, Blankenship RE, Sharples FP, Hiller RG, Birge RR, Frank HA. Spectroscopic properties of the main-form and high-salt peridinin–chlorophyll a proteins from Amphidinium carterae. Biochemistry. 2004;43:1478–1487. doi: 10.1021/bi0357964. [DOI] [PubMed] [Google Scholar]
- 21.Schulte T, Niedzwiedzki DM, Birge RR, Hiller RG, Polivka T, Hofmann E, Frank HA. Identification of a single peridinin sensing Chl-a excitation in reconstituted PCP by crystallography and spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20764–20769. doi: 10.1073/pnas.0908938106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharples FP, Wrench PM, Ou K, Hiller RG. Two distinct forms of the peridinin–chlorophyll a-protein from Amphidinium carterae. Biochim. Biophys. Acta. 1996;1276:117–123. doi: 10.1016/0005-2728(96)00066-7. [DOI] [PubMed] [Google Scholar]
- 23.Triplett EL, Jovine RV, Govind NS, Roman SJ, Chang SS, Prezelin BB. Characterization of two full-length cDNA sequences encoding for apoproteins of peridinin–chlorophyll a-protein (PCP) complexes. Mol. Mar. Biol. Biotechnol. 1993;2:246–254. [PubMed] [Google Scholar]
- 24.Reichman JR, Wilcox TP, Vize PD. PCP gene family in Symbiodinium from Hippopus hippopus: low levels of concerted evolution, isoform diversity, and spectral tuning of chromophores. Mol. Biol. Evol. 2003;20:2143–2154. doi: 10.1093/molbev/msg233. [DOI] [PubMed] [Google Scholar]
- 25.Carlos AA, Baillie BK, Kawachi M, Maruyama T. Phylogenetic position of Symbiodinium (Dinophyceae) isolates from tridacnids (Bivalvia), cardiids (Bivalvia), a sponge (Porifera), a soft coral (Anthozoa), and a free-living strain. J. Phycol. 1999;35:1054–1062. [Google Scholar]
- 26.Schaegger H. Tricine-SDS-PAGE, Nat. Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 27.Yue H, Kang Y, Zhang H, Gao X, Blankenship RE. Expression and characterization of the diheme cytochrome c subunit of the cytochrome bc complex in Heliobacterium modesticaldum. Arch. Biochem. Biophys. 2012;517:131–137. doi: 10.1016/j.abb.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Gao X, Xin Y, Bell PD, Wen J, Blankenship RE. Structural analysis of alternative complex III in the photosynthetic electron transfer chain of Chloroflexus aurantiacus. Biochemistry. 2010;49:6670–6679. doi: 10.1021/bi100858k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Huang RY, Jalili PR, Irungu JW, Nicol GR, Ray KB, Rohrs HW, Gross ML. Improved mass spectrometric characterization of protein glycosylation reveals unusual glycosylation of maize-derived bovine trypsin. Anal. Chem. 2010;82:10095–10101. doi: 10.1021/ac1020722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plumb RS, Johnson KA, Rainville P, Smith BW, Wilson ID, Castro-Perez JM, Nicholson JK. UPLC/MSE; a new approach for generating molecular fragment information for biomarker structure elucidation, Rapid Commun. Mass Spectrom. 2006;20:1989–1994. doi: 10.1002/rcm.2550. [DOI] [PubMed] [Google Scholar]
- 31.Zigmantas D, Polivka T, Hiller RG, Yartsev A, Sundstrom V. Spectroscopic and dynamic properties of the peridinin lowest singlet excited states. J. Phys. Chem. A. 2001;105:10296–10306. [Google Scholar]
- 32.Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations forassaying chlorophyll-a and chlorophyll-b extracted with 4 different solvents — verification of the concentration of chlorophyll standards by atomic-absorption spectroscopy. Biochim. Biophys. Acta. 1989;975:384–394. [Google Scholar]
- 33.Jeffrey SW, Haxo FT. Photosynthetic pigments of symbiotic dinoflagellates (zooxanthellae) from corals and clams. Biol. Bull. 1968;135:149–165. [Google Scholar]
- 34.Podsiadlo P, Michel M, Lee J, Verploegen E, Wong Shi Kam N, Ball V, Qi Y, Hart AJ, Hammond PT, Kotov NA. Exponential growth of LBL films with incorporated inorganic sheets. Nano Lett. 2008;8:1762–1770. doi: 10.1021/nl8011648. [DOI] [PubMed] [Google Scholar]
- 35.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 36.Xu H, Freitas MA. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinformatics. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu H, Zhang L, Freitas MA. Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix MS/MS search engine. J. Proteome Res. 2008;7:138–144. doi: 10.1021/pr070363z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nassoury N, Cappadocia M, Morse D. Plastid ultrastructure defines the protein import pathway in dinoflagellates. J. Cell Sci. 2003;116:2867–2874. doi: 10.1242/jcs.00517. [DOI] [PubMed] [Google Scholar]
- 39.Smeekens S, Bauerle C, Hageman J, Keegstra K, Weisbeek P. The role of the transit peptide in the routing of precursors toward different chloroplast compartments. Cell. 1986;46:365–375. doi: 10.1016/0092-8674(86)90657-4. [DOI] [PubMed] [Google Scholar]
- 40.Schnell DJ. Protein targeting to the thylakoid membrane. Annu. Rev. Plant Physiol. Plant. Mol. Biol. 1998;49:97–126. doi: 10.1146/annurev.arplant.49.1.97. [DOI] [PubMed] [Google Scholar]
- 41.Balsera M, Soll J, Bolter B. Protein import machineries in endosymbiotic organelles. Cell. Mol. Life Sci. 2009;66:1903–1923. doi: 10.1007/s00018-009-8644-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rawyler A, Meylan-Bettex M, Siegenthaler PA. Galactolipid export from envelope to thylakoid membranes in intact chloroplasts. II. A general process with a key role for the envelope in the establishment of lipid asymmetry in thylakoid membranes. Biochim. Biophys. Acta. 1995;1233:123–133. doi: 10.1016/0005-2736(94)00248-n. [DOI] [PubMed] [Google Scholar]
- 43.Schulte T, Hiller RG, Hofmann E. X-ray structures of the peridinin–chlorophyll-protein reconstituted with different chlorophylls. FEBS Lett. 2010;584:973–978. doi: 10.1016/j.febslet.2010.01.041. [DOI] [PubMed] [Google Scholar]
- 44.Polivka T, Hiller RG, Frank HA. Spectroscopy of the peridinin–chlorophyll-a protein: insight into light-harvesting strategy of marine algae. Arch. Biochem. Biophys. 2007;458:111–120. doi: 10.1016/j.abb.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 45.Schulte T, Johanning S, Hofmann E. Structure and function of native and refolded peridinin–chlorophyll-proteins from dinoflagellates. Eur. J. Cell Biol. 2010;89:990–997. doi: 10.1016/j.ejcb.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Huang D, Cramer WA. Stoichiometrically bound beta-carotene in the cytochrome b6f complex of oxygenic photosynthesis protects against oxygen damage. J. Biol. Chem. 1999;274:1581–1587. doi: 10.1074/jbc.274.3.1581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.