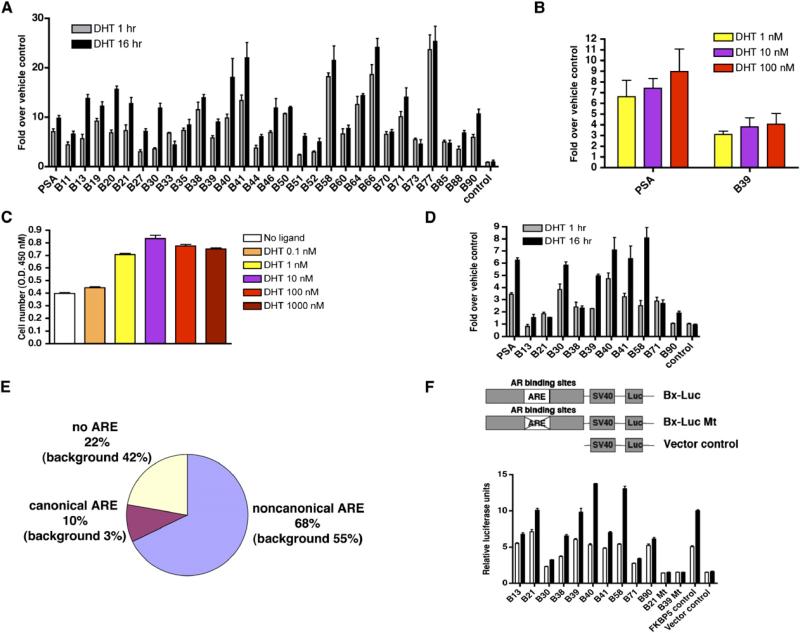
Figure 2. Characterization of AR Binding Sites on Chromosomes 21 and 22.
(A) Standard ChIP assays of AR recruitment to various potential AR binding regions. LNCaP cells were treated with 100 nM DHT for 1 or 16 hr. ChIP assays were performed with anti-AR antibodies. Immunoprecipitated DNA was quantified by real-time PCR using primers (Table S1) spanning various potential AR binding regions.
(B and C) Effects of DHT concentration on AR binding to chromatin and cell proliferation. (B) Androgen-depleted LNCaP cells were treated with 1, 10, or 100 nM DHT for 1 hr. ChIP assays were performed with an anti-AR antibody, and DNA precipitates were measured with real-time PCR using primers spanning the PSA enhancer and the B39 site. (C) LNCaP cells were cultured in hormone-depleted medium for 3 days and then treated with vehicle or DHT from 0.1 to 1000 nM for another 3 days. Cell proliferation was measured using the WST-1 assay.
(D) Standard ChIP assays of Pol II recruitment to various potential AR binding regions. ChIP assays were performed with anti-Pol II antibodies.
(E) Distribution of the types of AR binding motifs within AR binding sites and the chromosome 21 and 22 genomic background.
(F) AR binding regions function as enhancers. Ten selected AR binding regions, mutant AR binding regions with deleted AREs (B21 and B39), and the FKBP5 enhancer were subcloned into the pGL3-promoter vector and transfected into androgen-depleted LNCaP cells. Empty pGL3 promoter vector was used as a negative control. Cells were stimulated with 100 nM DHT or vehicle for 24 hr. The data were presented as the mean ± SE of two to three replicates, (A)–(D) and (F).