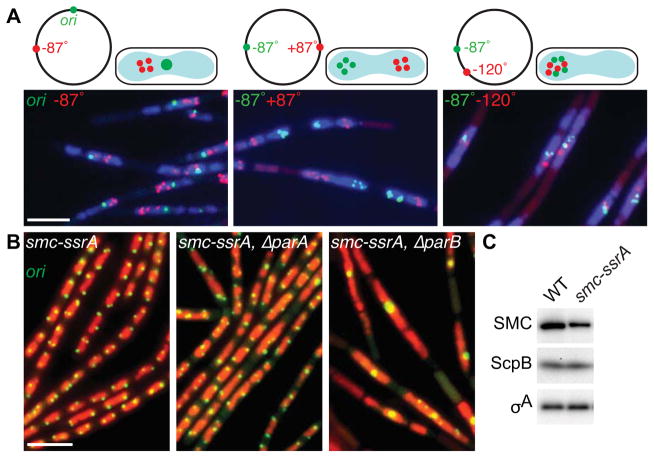
Figure 2. ParB-mediated recruitment of SMC promotes efficient chromosome segregation.
(A) Bulk chromosome segregation is blocked upon SMC inactivation. Representative images of DAPI-stained nucleoids (blue) and indicated chromosomal loci (green and red) in the smcts mutant (BWX2378, BWX2116, BWX2110) grown in CH medium for 1.5 h after shifting to 42°C. Schematic representations of the loci analyzed and their subcellular locations are shown above the micrographs. 72–76% (n>1320 per strain) had nucleoids and foci similar to those shown in (A). (B) Origin segregation is impaired in ParB mutants when SMC levels are reduced. Representative micrographs of DAPI-stained nucleoids (false-colored red) and origins (green) in cells (BWX1497, left panel) harboring an smc-ssrA fusion that results in a 2.5-fold reduction in SMC levels. The cells in the middle panel (BWX2551) harbor an in-frame deletion of parA and those in the right panel (BWX1569) lack parB. 2% (n=1165) of smc-ssrA cells; 8% (n=1038) of ΔparA, smc-ssrA cells; and 34% (n=1214) of ΔparB, smc-ssrA cells had a single bright origin focus. 5%, 13%, and 80%, respectively had an unsegregated nucleoid. Representative images of ParA and ParB mutants that have wild-type levels of SMC and quantitative analysis of nucleoid size in all strains can be found in Figure S2D. Scale bars are 4 μm. (C) Immunoblot analysis of SMC, ScpB, and a loading control (σA) in wild-type and the smc-ssrA mutant under conditions in which the adaptor protein is not induced. See also Figure S2.